O43318
Gene name |
MAP3K7 (TAK1) |
Protein name |
Mitogen-activated protein kinase kinase kinase 7 |
Names |
EC 2.7.11.25 , Transforming growth factor-beta-activated kinase 1 , TGF-beta-activated kinase 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:6885 |
EC number |
2.7.11.25: Protein-serine/threonine kinases |
Protein Class |
|
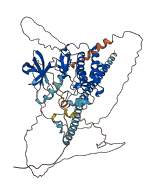
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
36-291 (Protein kinase domain) |
Relief mechanism |
Ligand binding, Partner binding |
Assay |
|
Accessory elements
174-194 (Activation loop from InterPro)
Target domain |
36-291 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
174-194 (Activation loop from InterPro)
Target domain |
36-291 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

24 structures for O43318
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2EVA | X-ray | 200 A | A | 31-303 | PDB |
| 2YIY | X-ray | 249 A | A | 31-303 | PDB |
| 4GS6 | X-ray | 220 A | A | 31-303 | PDB |
| 4L3P | X-ray | 268 A | A | 31-303 | PDB |
| 4L52 | X-ray | 254 A | A | 31-303 | PDB |
| 4L53 | X-ray | 255 A | A | 31-303 | PDB |
| 4O91 | X-ray | 239 A | A | 31-303 | PDB |
| 5E7R | X-ray | 211 A | A | 31-303 | PDB |
| 5GJD | X-ray | 279 A | A | 31-303 | PDB |
| 5GJF | X-ray | 289 A | A | 31-303 | PDB |
| 5GJG | X-ray | 261 A | A | 31-303 | PDB |
| 5J7S | X-ray | 237 A | A | 31-303 | PDB |
| 5J8I | X-ray | 240 A | A | 31-303 | PDB |
| 5J9L | X-ray | 275 A | A | 31-303 | PDB |
| 5JGA | X-ray | 200 A | A | 31-303 | PDB |
| 5JGB | X-ray | 280 A | A | 31-303 | PDB |
| 5JGD | X-ray | 310 A | A | 31-303 | PDB |
| 5JH6 | X-ray | 237 A | A | 31-303 | PDB |
| 5JK3 | X-ray | 237 A | A | 31-303 | PDB |
| 5V5N | X-ray | 201 A | A | 31-303 | PDB |
| 7NTH | X-ray | 197 A | A | 31-303 | PDB |
| 7NTI | X-ray | 198 A | A | 31-303 | PDB |
| 8GW3 | X-ray | 205 A | A/B/C/D | 15-303 | PDB |
| AF-O43318-F1 | Predicted | AlphaFoldDB |
341 variants for O43318
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV002543632 RCV001313769 CA3928785 rs139666795 |
9 | S>F | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
rs1582233787 RCV000845245 |
42 | V>missing | Cardiospondylocarpofacial syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs886039234 RCV000254560 |
44 | R>missing | Cardiospondylocarpofacial syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1776911945 RCV001194646 |
48 | G>E | Cardiospondylocarpofacial syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000254567 RCV001387545 VAR_077341 rs886039236 |
50 | V>missing | Cardiospondylocarpofacial syndrome CSCF [ClinVar, UniProt] | Yes |
ClinVar UniProt dbSNP |
|
VAR_077341 rs886039236 |
50 | V>del | CSCF [UniProt] | Yes |
UniProt dbSNP |
|
rs886039231 VAR_077342 CA10588007 RCV000254559 |
70 | E>Q | Frontometaphyseal dysplasia 2 FMD2 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA143017447 RCV000850358 rs929527043 |
83 | R>H | Cardiospondylocarpofacial syndrome [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP |
|
VAR_077343 RCV000254562 rs886039232 CA10588008 |
100 | V>E | Frontometaphyseal dysplasia 2 FMD2 [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001859473 CA10588011 VAR_077344 RCV000254563 rs886039235 |
110 | G>C | Cardiospondylocarpofacial syndrome CSCF [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
CA10588009 rs886039233 VAR_077345 RCV000254566 |
168 | G>R | Frontometaphyseal dysplasia 2 FMD2; increases autophosphorylation; no effect on MAPK signaling; no effect on NF-kappa-B signaling [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001266455 rs1776343243 |
191 | G>E | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1776245887 RCV001281689 |
211 | D>G | Cardiospondylocarpofacial syndrome [ClinVar] | Yes |
ClinVar dbSNP |
|
rs886039237 RCV000254561 CA10588013 VAR_077346 |
241 | W>R | Cardiospondylocarpofacial syndrome CSCF [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs767139595 CA365010309 RCV000985011 |
329 | N>I | MAP3K7-related disorder [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV001270770 rs1775677189 |
451 | G>S | Frontometaphyseal dysplasia 2 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA10588006 RCV000254565 VAR_077347 RCV001530168 rs886039230 |
512 | P>L | Frontometaphyseal dysplasia 2 FMD2; does not affect interaction with TAB2; does not affect homodimerization; increases autophosphorylation; increases MAPK signaling; increases NF-kappa-B signaling [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt dbSNP gnomAD |
|
rs752301948 CA3928791 |
3 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752301948 CA365021164 |
3 | T>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3928790 rs370126466 |
6 | A>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs754729477 CA3928789 |
7 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA3928787 rs766181895 |
8 | S>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs552731765 CA3928786 |
8 | S>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1435177696 CA365021063 |
10 | S>F | No |
ClinGen TOPMed |
|
|
rs1051442933 CA143029513 |
12 | S>P | No |
ClinGen gnomAD |
|
|
rs775701255 CA3928779 |
13 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1452414465 CA365021009 |
14 | S>L | No |
ClinGen gnomAD |
|
|
RCV001349323 rs772116030 CA3928778 |
15 | A>S | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs879577986 CA143029484 |
15 | A>V | No |
ClinGen gnomAD |
|
|
rs771456548 CA3928775 |
17 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs778123770 CA3928773 |
19 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365020931 rs1181635158 |
19 | I>T | No |
ClinGen gnomAD |
|
|
rs532622360 CA3928774 |
19 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1243505437 CA365020923 |
20 | E>Q | No |
ClinGen gnomAD |
|
|
rs1201891278 CA365020908 |
21 | A>T | No |
ClinGen gnomAD |
|
|
CA365020891 rs1259724512 |
22 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
CA3928772 rs770391295 |
23 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747607026 CA3928771 |
24 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA3928770 rs780760405 |
25 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA143029453 rs936804341 |
27 | N>S | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 30 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs750287415 CA3928766 |
32 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA3928767 rs779720104 |
32 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs765077005 CA3928764 |
33 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
rs1363754782 CA365020692 |
34 | K>R | No |
ClinGen gnomAD |
|
|
CA365020694 rs1363754782 |
34 | K>T | No |
ClinGen gnomAD |
|
|
CA365018265 rs1395596489 |
51 | C>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA143020071 rs77759048 |
55 | W>R | No |
ClinGen Ensembl |
|
|
rs751751419 CA3928730 |
56 | R>S | No |
ClinGen ExAC gnomAD |
|
|
CA365018107 rs766705070 |
57 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA3928729 rs766705070 |
57 | A>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 57 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 66 | E>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs773394242 CA3928727 |
73 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs773394242 CA3928726 |
73 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs762124152 CA3928724 |
76 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1582233598 CA365017878 RCV000792551 |
77 | E>A | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs758501507 CA3928709 |
79 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
rs973658423 CA143017440 |
85 | N>D | No |
ClinGen TOPMed |
|
|
rs765425385 CA3928707 |
85 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA365017472 rs1204627384 |
86 | H>D | No |
ClinGen gnomAD |
|
|
rs369978696 CA143017427 |
87 | P>A | No |
ClinGen ESP ExAC gnomAD |
|
|
rs764513500 CA3928704 |
87 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs369978696 CA3928705 |
87 | P>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA gnomAD |
|
rs369978696 CA365017461 |
87 | P>T | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1309935275 CA365017419 |
91 | K>Q | No |
ClinGen gnomAD |
|
|
CA143017401 rs959469426 |
98 | N>S | No |
ClinGen Ensembl |
|
|
CA365017324 rs1314103804 |
99 | P>L | No |
ClinGen gnomAD |
|
|
CA143011466 rs948221759 |
100 | V>L | No |
ClinGen TOPMed |
|
|
rs922842241 CA143011463 |
101 | C>S | No |
ClinGen Ensembl |
|
|
CA365016068 rs1340670657 |
106 | Y>* | No |
ClinGen gnomAD |
|
|
rs1064795771 CA16618340 RCV000478415 |
107 | A>T | No |
ClinGen ClinVar Ensembl dbSNP |
|
| TCGA novel | 111 | S>F | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 115 | V>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1221277357 CA365015681 |
117 | H>D | No |
ClinGen gnomAD |
|
|
CA365015601 rs1322286349 |
121 | P>Q | No |
ClinGen gnomAD |
|
|
CA3928659 rs772970352 |
123 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA143009930 rs372169648 |
124 | Y>C | No |
ClinGen ESP |
|
|
rs1055343222 CA143009928 |
126 | T>A | No |
ClinGen TOPMed |
|
|
rs1402882263 CA365015485 |
128 | A>D | No |
ClinGen gnomAD |
|
|
rs139832891 CA3928657 |
130 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs776294688 CA3928656 |
131 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1253556441 CA365015412 |
132 | S>N | No |
ClinGen TOPMed |
|
|
rs866711961 CA143009897 |
138 | S>F | No |
ClinGen Ensembl |
|
|
CA143009887 rs34261934 |
139 | Q>H | No |
ClinGen Ensembl |
|
|
rs1417350892 CA365015282 |
139 | Q>R | No |
ClinGen gnomAD |
|
|
CA365015262 rs1405746578 |
140 | G>E | No |
ClinGen gnomAD |
|
| TCGA novel | 146 | S>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1216965396 CA365015115 |
149 | P>L | No |
ClinGen gnomAD |
|
|
rs1265335108 CA365015084 |
151 | A>T | No |
ClinGen TOPMed |
|
| TCGA novel | 155 | R>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 162 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365014028 rs943102765 |
167 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
rs943102765 CA143007116 |
167 | G>W | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 169 | T>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1057517758 RCV000414538 CA16042655 |
174 | C>Y | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA3928616 rs199879592 |
178 | T>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA143007098 rs868774819 |
180 | C>R | No |
ClinGen Ensembl |
|
| TCGA novel | 184 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 187 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3928614 rs760377087 |
188 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA365012710 rs1439524424 |
203 | G>V | No |
ClinGen gnomAD |
|
|
CA365012704 rs1200838990 |
204 | S>C | No |
ClinGen gnomAD |
|
|
rs1460160552 CA365012699 |
204 | S>I | No |
ClinGen gnomAD |
|
|
rs1460160552 CA365012698 |
204 | S>N | No |
ClinGen gnomAD |
|
|
CA365012679 rs1201135517 |
205 | N>K | No |
ClinGen gnomAD |
|
|
CA365012662 rs1554184177 RCV000523069 COSM1634986 COSM1634985 |
206 | Y>C | liver [Cosmic] | No |
ClinGen cosmic curated ClinVar Ensembl dbSNP |
| TCGA novel | 209 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA143004424 rs549128142 |
209 | K>T | No |
ClinGen gnomAD |
|
|
CA3928584 rs760970094 |
212 | V>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 212 | V>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3928583 rs775953358 |
216 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1448649376 CA365012495 |
217 | I>V | No |
ClinGen gnomAD |
|
|
rs1338362778 CA365012458 |
219 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA3928582 rs772534247 |
220 | W>* | No |
ClinGen ExAC gnomAD |
|
|
CA143004396 rs772534247 |
220 | W>C | No |
ClinGen ExAC gnomAD |
|
|
CA3928581 rs746282865 |
224 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1157398284 COSM1082640 CA365012385 COSM1596540 |
225 | R>C | endometrium Variant assessed as Somatic; 4.619e-05 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA143004349 rs200376548 |
225 | R>H | No |
ClinGen Ensembl |
|
| TCGA novel | 226 | R>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3928578 rs748681814 |
232 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA365012277 rs1582204926 |
233 | G>S | No |
ClinGen Ensembl |
|
|
rs755674001 CA3928576 |
234 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA16618339 RCV000479656 rs886039237 |
241 | W>G | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA365012149 rs1188476695 |
242 | A>T | No |
ClinGen gnomAD |
|
|
CA143004332 rs879103527 |
244 | H>R | No |
ClinGen Ensembl |
|
|
COSM3831241 COSM3831240 CA143003140 rs976691872 |
248 | R>* | breast [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA365011974 rs1562095491 |
250 | P>S | No |
ClinGen Ensembl |
|
| TCGA novel | 252 | I>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365011959 rs1358054091 |
252 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs149444442 CA3928560 |
259 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA365011905 rs1340569337 |
260 | E>G | No |
ClinGen gnomAD |
|
|
rs772558226 CA3928559 |
260 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA365011897 rs1434933903 |
261 | S>T | No |
ClinGen gnomAD |
|
|
rs780812265 CA3928556 |
262 | L>P | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 262 | L>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA3928555 rs754695346 |
263 | M>K | No |
ClinGen ExAC gnomAD |
|
|
CA365011873 rs1421803205 |
265 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1421803205 CA365011874 |
265 | R>G | No |
ClinGen gnomAD |
|
|
COSM1082638 rs746745976 CA3928554 COSM1596542 |
265 | R>H | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA143003108 rs978403563 |
268 | S>C | No |
ClinGen Ensembl |
|
|
rs1478218204 CA365011838 |
268 | S>P | No |
ClinGen gnomAD |
|
|
CA3928553 rs138091630 |
273 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3928552 COSM3831239 rs758224984 COSM3831238 |
274 | R>H | Variant assessed as Somatic; 0.0 impact. breast [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA3928551 rs750338877 |
275 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3928549 rs756248428 |
277 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 279 | E>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs752889791 CA3928548 |
280 | I>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 283 | I>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365011631 rs1197604380 |
284 | M>I | No |
ClinGen TOPMed gnomAD |
|
|
rs201660449 CA3928547 |
284 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1355612914 CA365011622 |
285 | T>S | No |
ClinGen TOPMed |
|
|
rs759853469 CA3928546 |
288 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA3928545 rs199615272 |
289 | R>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs781446047 CA3928530 |
292 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA3928529 rs755212534 |
293 | G>E | No |
ClinGen ExAC gnomAD |
|
|
rs1017533232 CA143001985 |
294 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1017533232 CA365010680 |
294 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1359798338 CA365010677 |
294 | A>V | No |
ClinGen gnomAD |
|
|
CA365010667 rs1409320544 |
296 | E>K | No |
ClinGen TOPMed |
|
|
CA3928527 rs766563050 |
308 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs1165240514 CA365010549 |
308 | G>R | No |
ClinGen TOPMed |
|
|
CA143001981 rs1008855643 |
310 | S>N | No |
ClinGen TOPMed |
|
|
CA3928526 rs763367393 |
313 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA3928524 rs765667865 |
315 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs762328133 CA3928523 |
316 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA365010474 rs1471178063 |
318 | S>P | No |
ClinGen TOPMed |
|
|
CA365010454 rs1217973285 |
319 | F>C | No |
ClinGen gnomAD |
|
|
CA365010438 rs1487091258 |
320 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1244444578 CA365010418 |
321 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs754318578 CA3928505 |
321 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA3928504 rs536467087 |
322 | I>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA3928503 rs760227252 |
327 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA365010330 rs1213420677 |
328 | S>N | No |
ClinGen TOPMed |
|
|
rs767139595 CA3928501 |
329 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365010296 rs1325197630 |
330 | K>R | No |
ClinGen TOPMed |
|
|
rs1262255199 CA365010248 |
333 | T>I | No |
ClinGen TOPMed |
|
|
rs374740115 CA143000814 |
333 | T>S | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1582197017 CA365010215 |
335 | M>R | No |
ClinGen Ensembl |
|
|
CA3928499 rs774158291 |
337 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
CA143000784 rs890285458 |
338 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
CA365010168 rs1457177066 |
338 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA365010166 rs1457177066 |
338 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA365010150 rs1456500923 |
339 | P>S | No |
ClinGen gnomAD |
|
|
rs749145408 CA3928497 |
342 | N>D | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA365010072 rs1476587917 |
343 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA3928495 rs769922850 |
345 | I>T | No |
ClinGen ExAC gnomAD |
|
|
COSM1187116 rs773273184 COSM1187115 CA3928496 |
345 | I>V | lung [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
rs747046300 CA3928494 |
347 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1185961784 CA365010032 |
347 | R>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 349 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs371531755 CA3928493 |
349 | E>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs758604367 CA143000765 |
352 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs1317447514 CA365009942 |
353 | L>S | No |
ClinGen gnomAD |
|
|
rs931867674 CA365009881 |
355 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs200449319 CA3928491 |
357 | A>G | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1582196811 CA365009843 |
359 | Q>K | No |
ClinGen Ensembl |
|
| TCGA novel | 360 | Q>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs776668005 CA3928473 |
361 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1331566897 CA365008970 |
365 | R>C | No |
ClinGen gnomAD |
|
|
CA3928472 rs772019658 |
365 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365008969 rs772019658 |
365 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3928470 rs542958423 |
367 | S>N | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1406540048 CA365008905 |
369 | G>R | No |
ClinGen TOPMed |
|
|
CA3928468 rs749595493 |
370 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA3928469 rs757475227 |
370 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365008850 rs778131562 |
372 | R>C | No |
ClinGen ExAC gnomAD |
|
|
CA3928467 rs778131562 |
372 | R>G | No |
ClinGen ExAC gnomAD |
|
|
CA143000215 rs961650989 |
372 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA3928466 rs756606414 |
374 | S>N | No |
ClinGen ExAC gnomAD |
|
|
CA3928465 rs368244160 |
375 | S>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3928464 rs781747384 |
377 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs751184932 CA365008720 |
379 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365008715 rs1282326742 |
380 | P>S | No |
ClinGen gnomAD |
|
|
CA3928460 rs762711020 |
382 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365008659 rs1279831807 |
385 | G>A | No |
ClinGen gnomAD |
|
|
rs1160244930 CA365008665 |
385 | G>C | No |
ClinGen gnomAD |
|
|
CA3928457 rs375763475 |
387 | R>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1377832888 CA365008633 |
388 | M>V | No |
ClinGen gnomAD |
|
|
CA3928455 rs759545618 |
389 | S>C | No |
ClinGen ExAC gnomAD |
|
|
CA3928454 rs759545618 |
389 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs1381308365 CA365008590 |
391 | D>Y | No |
ClinGen gnomAD |
|
|
rs778011461 COSM1739297 CA365008574 COSM1739298 |
392 | M>K | haematopoietic_and_lymphoid_tissue [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs778011461 CA3928450 RCV001300164 |
392 | M>R | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs778011461 CA3928451 |
392 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771091309 CA3928452 |
392 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1201119850 CA365008542 |
395 | I>V | No |
ClinGen TOPMed |
|
|
CA3928448 rs748605104 |
399 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA3928447 rs781547814 |
400 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365008494 rs1214345872 |
400 | A>V | No |
ClinGen gnomAD |
|
|
CA3928445 rs751058214 |
401 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA3928444 rs779736807 |
403 | T>A | No |
ClinGen ExAC |
|
| TCGA novel | 404 | A>= | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM484457 CA3928418 rs752610893 |
404 | A>V | kidney [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA3928417 rs767502781 |
406 | S>A | No |
ClinGen ExAC gnomAD |
|
|
rs1258442093 CA365008450 |
406 | S>Y | No |
ClinGen TOPMed |
|
|
rs763016638 CA3928416 |
408 | P>A | No |
ClinGen ExAC gnomAD |
|
|
CA365008439 rs763016638 |
408 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1307538507 CA365008433 |
409 | K>E | No |
ClinGen gnomAD |
|
|
VAR_080761 CA3928415 rs201721045 |
410 | R>Q | found in a consanguineous family with intellectual disability; unknown pathological significance [UniProt] | No |
ClinGen UniProt ExAC dbSNP gnomAD |
|
rs1040962323 CA142998231 |
410 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA365008420 RCV001057789 rs1426464104 |
411 | G>D | No |
ClinGen ClinVar TOPMed dbSNP |
|
|
rs762121500 COSM1446472 RCV001350590 CA3928413 |
413 | R>C | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ClinVar ExAC NCI-TCGA TOPMed dbSNP gnomAD |
|
CA3928412 rs776824877 |
413 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3928409 rs776133610 |
419 | G>D | No |
ClinGen ExAC gnomAD |
|
|
CA365008362 rs1467091855 |
420 | N>S | No |
ClinGen TOPMed |
|
|
rs142816839 CA3928407 |
421 | I>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3928408 rs772708153 |
421 | I>T | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 421 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA142998179 rs891888303 |
423 | D>H | No |
ClinGen TOPMed |
|
|
CA365008347 rs891888303 |
423 | D>Y | No |
ClinGen TOPMed |
|
|
rs1398765066 CA365008333 |
425 | P>S | No |
ClinGen gnomAD |
|
|
rs138804542 CA3928406 |
426 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA3928405 rs147070999 |
428 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA3928404 rs748938590 |
429 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs777451903 CA3928403 |
430 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs772656967 CA3928388 |
431 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM1082634 rs774760752 CA3928386 COSM1596545 |
433 | G>R | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
rs1216888405 CA365007051 |
436 | R>T | No |
ClinGen gnomAD |
|
|
CA3928385 rs770452435 |
437 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs537649538 CA3928384 |
437 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA3928383 rs568609705 |
440 | I>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1279939521 CA365007012 |
442 | D>G | No |
ClinGen gnomAD |
|
|
CA365007004 rs1356157105 |
443 | L>S | No |
ClinGen gnomAD |
|
|
CA365006995 rs1293903060 |
445 | V>I | No |
ClinGen gnomAD |
|
|
rs1416547471 CA365006974 |
448 | T>I | No |
ClinGen gnomAD |
|
|
rs1280748238 CA365006958 |
450 | P>L | No |
ClinGen gnomAD |
|
|
CA142992893 rs1031799928 |
452 | Q>L | No |
ClinGen Ensembl |
|
|
rs1344447752 CA365004627 |
454 | S>N | No |
ClinGen gnomAD |
|
|
rs1449822155 CA365004593 |
459 | S>T | No |
ClinGen TOPMed |
|
|
rs1194502439 CA365004586 |
460 | P>H | No |
ClinGen gnomAD |
|
|
rs774909552 CA3928366 |
461 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs771408689 CA3928365 |
463 | R>K | No |
ClinGen ExAC gnomAD |
|
|
rs1193330160 CA365004543 |
466 | T>I | No |
ClinGen gnomAD |
|
|
CA3928364 rs762460576 |
467 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3928361 rs747832498 |
469 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1003317029 CA142986043 |
470 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs768479279 CA3928359 |
471 | T>S | No |
ClinGen ExAC gnomAD |
|
|
CA3928358 rs746842603 |
472 | S>L | No |
ClinGen ExAC gnomAD |
|
|
CA365004488 rs1582161840 |
476 | T>A | No |
ClinGen Ensembl |
|
|
rs1452061275 CA365004482 |
477 | R>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs183005565 CA3928356 |
477 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA365004474 rs1327472475 |
478 | S>I | No |
ClinGen TOPMed gnomAD |
|
|
CA365004476 rs1327472475 |
478 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1562078333 CA365004467 |
479 | H>R | No |
ClinGen Ensembl |
|
|
rs375936720 CA3928355 |
480 | P>A | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1005730293 CA142986001 |
481 | W>R | No |
ClinGen Ensembl |
|
|
CA365004440 rs1305343134 |
483 | P>R | No |
ClinGen TOPMed |
|
|
rs756301309 CA3928353 |
486 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1364553569 CA365016308 |
489 | T>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA3928332 COSM94667 rs375179983 |
490 | N>S | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs751903983 CA3928331 |
492 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1255385324 CA365016208 |
493 | D>E | No |
ClinGen gnomAD |
|
|
rs1210065874 CA365016149 |
496 | I>L | No |
ClinGen gnomAD |
|
|
rs750911139 CA3928328 |
498 | M>V | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 499 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA143021235 rs904380452 |
502 | T>A | No |
ClinGen Ensembl |
|
|
CA365015944 rs1279741295 |
504 | D>G | No |
ClinGen gnomAD |
|
|
rs1240623710 CA365015929 |
505 | H>Y | No |
ClinGen gnomAD |
|
|
CA365015120 rs886039230 |
512 | P>Q | No |
ClinGen gnomAD |
|
|
CA365015118 rs886039230 |
512 | P>R | No |
ClinGen gnomAD |
|
|
CA365015094 rs1430533332 |
513 | C>Y | No |
ClinGen gnomAD |
|
|
rs750857862 CA3928310 |
515 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA3928309 rs765671703 |
516 | S>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA143020675 rs955925138 |
519 | S>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
| TCGA novel | 520 | M>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365014854 rs1460084200 |
525 | Q>R | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs757780457 CA3928308 |
532 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA365014749 rs1562074791 |
536 | V>A | No |
ClinGen Ensembl |
|
| TCGA novel | 539 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA365014645 rs1166077451 |
545 | Q>R | No |
ClinGen gnomAD |
|
|
rs1474999375 CA365014613 |
547 | K>R | No |
ClinGen gnomAD |
|
|
CA3928289 rs779276833 |
556 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778400307 CA3928286 |
564 | T>A | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs755574294 CA3928285 |
564 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA365014265 rs1358908852 |
565 | S>P | No |
ClinGen TOPMed |
|
|
rs767221108 CA3928284 |
566 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA3928283 rs767221108 |
566 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1457817469 CA365014250 |
568 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA365014249 rs1457817469 |
568 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1365432543 CA365014240 |
569 | Q>P | No |
ClinGen TOPMed |
|
|
rs766248028 CA3928280 |
571 | H>L | No |
ClinGen ExAC gnomAD |
|
|
rs762857691 CA3928278 |
575 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs1304532901 CA365014187 |
576 | D>E | No |
ClinGen gnomAD |
|
| TCGA novel | 577 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1236776312 CA365014173 |
578 | N>S | No |
ClinGen gnomAD |
|
|
CA143019406 rs193156060 |
579 | K>R | No |
ClinGen 1000Genomes gnomAD |
|
|
rs955602310 CA143019405 |
580 | S>R | No |
ClinGen TOPMed |
|
|
rs149129500 CA3928276 |
586 | Q>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1582152078 CA365014115 |
586 | Q>H | No |
ClinGen Ensembl |
|
|
rs760608455 CA3928275 |
588 | C>R | No |
ClinGen ExAC gnomAD |
|
|
rs1309758104 CA365014086 |
589 | K>R | No |
ClinGen gnomAD |
|
|
rs1432698401 CA365014066 |
590 | K>N | No |
ClinGen gnomAD |
|
| TCGA novel | 591 | Q>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 591 | Q>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs775565610 CA3928274 |
591 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746056485 CA3928272 |
594 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA365013974 rs1470177652 |
598 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
CA3928270 rs771627063 |
602 | R>* | No |
ClinGen ExAC gnomAD |
|
|
CA143019327 rs779104023 |
602 | R>Q | No |
ClinGen gnomAD |
|
|
CA365013901 rs1217645013 |
603 | Q>L | No |
ClinGen TOPMed |
No associated diseases with O43318
13 regional properties for O43318
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Receptor L-domain | 57 - 167 | IPR000494-1 |
| domain | Receptor L-domain | 361 - 480 | IPR000494-2 |
| domain | Protein kinase domain | 712 - 979 | IPR000719 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 714 - 966 | IPR001245 |
| domain | Furin-like cysteine-rich domain | 185 - 338 | IPR006211 |
| repeat | Furin-like repeat | 228 - 274 | IPR006212-1 |
| repeat | Furin-like repeat | 496 - 601 | IPR006212-2 |
| repeat | Furin-like repeat | 614 - 652 | IPR006212-3 |
| active_site | Tyrosine-protein kinase, active site | 833 - 845 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 718 - 745 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 712 - 968 | IPR020635 |
| domain | Growth factor receptor domain 4 | 505 - 636 | IPR032778 |
| domain | Epidermal growth factor receptor-like, transmembrane-juxtamembrane segment | 646 - 681 | IPR049328 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.25 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
7 GO annotations of cellular component
| Name | Definition |
|---|---|
| ATAC complex | A chromatin remodelling complex that regulates transcription via acetylation primarily of nucleosomal histones H3 and possibly H4. Shares the histone acetylation (HAT) module of GCN5/PCAF-ADA2-ADA3-SGF29 (or orthologs) with the related SAGA complex (GO:0000124). Contains HAT subunits GCN5 or PCAF in a mutually exclusive manner. In addition to the HAT module contains DR1/NC2B, KAT14, MBIP, WDR5, YEATS2 and ZZZ3 or orthologs. Also regulates the activity of non-histone targets and orchestrates mitotic progression by regulating Cyclin A degradation through acetylation. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum membrane | The lipid bilayer surrounding the endoplasmic reticulum. |
| endosome membrane | The lipid bilayer surrounding an endosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
17 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| DNA-binding transcription factor binding | Binding to a DNA-binding transcription factor, a protein that interacts with a specific DNA sequence (sometimes referred to as a motif) within the regulatory region of a gene to modulate transcription. |
| histone kinase activity | Catalysis of the transfer of a phosphate group to a histone. |
| identical protein binding | Binding to an identical protein or proteins. |
| linear polyubiquitin binding | Binding to a linear polymer of ubiquitin. Linear ubiquitin polymers are formed by linking the amino-terminal methionine (M1) of one ubiquitin molecule to the carboxy-terminal glycine (G76) of the next. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| MAP kinase activity | Catalysis of the reaction |
| MAP kinase kinase kinase activity | Catalysis of the phosphorylation and activation of a MAP kinase kinase; each MAP kinase kinase can be phosphorylated by any of several MAP kinase kinase kinases. |
| MAP kinase kinase kinase kinase activity | Catalysis of the phosphorylation of serine and threonine residues in a mitogen-activated protein kinase kinase kinase (MAPKKK), resulting in activation of MAPKKK. MAPKKK signaling pathways relay, amplify and integrate signals from the plasma membrane to the nucleus in response to a diverse range of extracellular stimuli. |
| protein serine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase activity | Catalysis of the reactions |
| protein serine/threonine kinase binding | Binding to a protein serine/threonine kinase. |
| receptor tyrosine kinase binding | Binding to a receptor that possesses protein tyrosine kinase activity. |
| scaffold protein binding | Binding to a scaffold protein. Scaffold proteins are crucial regulators of many key signaling pathways. Although not strictly defined in function, they are known to interact and/or bind with multiple members of a signaling pathway, tethering them into complexes. |
| transcription coactivator binding | Binding to a transcription coactivator, a protein involved in positive regulation of transcription via protein-protein interactions with transcription factors and other proteins that positively regulate transcription. Transcription coactivators do not bind DNA directly, but rather mediate protein-protein interactions between activating transcription factors and the basal transcription machinery. |
| type II transforming growth factor beta receptor binding | Binding to a type II transforming growth factor beta receptor. |
| ubiquitin protein ligase binding | Binding to a ubiquitin protein ligase enzyme, any of the E3 proteins. |
38 GO annotations of biological process
| Name | Definition |
|---|---|
| activation of NF-kappaB-inducing kinase activity | The stimulation of the activity of NF-kappaB-inducing kinase through phosphorylation at specific residues. |
| anoikis | Apoptosis triggered by inadequate or inappropriate adherence to substrate e.g. after disruption of the interactions between normal epithelial cells and the extracellular matrix. |
| canonical NF-kappaB signal transduction | The process in which a signal is passed on to downstream components within the cell through the I-kappaB-kinase (IKK)-dependent activation of NF-kappaB. The cascade begins with activation of a trimeric IKK complex (consisting of catalytic kinase subunits IKKalpha and/or IKKbeta, and the regulatory scaffold protein NEMO) and ends with the regulation of transcription of target genes by NF-kappaB. In a resting state, NF-kappaB dimers are bound to I-kappaB proteins, sequestering NF-kappaB in the cytoplasm. Phosphorylation of I-kappaB targets I-kappaB for ubiquitination and proteasomal degradation, thus releasing the NF-kappaB dimers, which can translocate to the nucleus to bind DNA and regulate transcription. |
| cellular response to angiotensin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an angiotensin stimulus. Angiotensin is any of three physiologically active peptides (angiotensin II, III, or IV) processed from angiotensinogen. |
| cellular response to hypoxia | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating lowered oxygen tension. Hypoxia, defined as a decline in O2 levels below normoxic levels of 20.8 - 20.95%, results in metabolic adaptation at both the cellular and organismal level. |
| cellular response to tumor necrosis factor | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a tumor necrosis factor stimulus. |
| cytoplasmic pattern recognition receptor signaling pathway | The series of molecular signals initiated by the binding of a ligand from another organism to a cytosolic pattern recognition receptor (PRR). PRRs bind pathogen-associated molecular pattern (PAMPs), structures conserved among microbial species. |
| defense response to bacterium | Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism. |
| Fc-epsilon receptor signaling pathway | The series of molecular signals initiated by the binding of the Fc portion of immunoglobulin E (IgE) to an Fc-epsilon receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. The Fc portion of an immunoglobulin is its C-terminal constant region. |
| I-kappaB phosphorylation | The process of introducing a phosphate group into an inhibitor of kappa B (I-kappaB) protein. Phosphorylation of I-kappaB targets I-kappaB for ubiquitination and proteasomal degradation, thus releasing bound NF-kappaB dimers, which can translocate to the nucleus to bind DNA and regulate transcription. |
| immune response | Any immune system process that functions in the calibrated response of an organism to a potential internal or invasive threat. |
| inflammatory response | The immediate defensive reaction (by vertebrate tissue) to infection or injury caused by chemical or physical agents. The process is characterized by local vasodilation, extravasation of plasma into intercellular spaces and accumulation of white blood cells and macrophages. |
| interleukin-1-mediated signaling pathway | The series of molecular signals initiated by interleukin-1 binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| interleukin-17A-mediated signaling pathway | The series of molecular signals initiated by interleukin-17A binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| interleukin-33-mediated signaling pathway | The series of molecular signals initiated by interleukin-33 binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| JNK cascade | An intracellular protein kinase cascade containing at least a JNK (a MAPK), a JNKK (a MAPKK) and a JUN3K (a MAP3K). The cascade can also contain an additional tier |
| MAPK cascade | An intracellular protein kinase cascade containing at least a MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tiers |
| MyD88-dependent toll-like receptor signaling pathway | A toll-like receptor signaling pathway in which the MyD88 adaptor molecule mediates transduction of the signal. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
| negative regulation of gene expression | Any process that decreases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| nucleotide-binding domain, leucine rich repeat containing receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a nucleotide-binding domain, leucine rich repeat containing receptor (NLR), and ending with the regulation of a downstream cellular process. NLRs are cytoplasmic receptors defined by their tripartite domain architecture that contains |
| p38MAPK cascade | An intracellular protein kinase cascade containing at least a p38 MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tier |
| positive regulation of canonical NF-kappaB signal transduction | Any process that activates or increases the frequency, rate or extent of I-kappaB kinase/NF-kappaB signaling. |
| positive regulation of cell cycle | Any process that activates or increases the rate or extent of progression through the cell cycle. |
| positive regulation of cell size | Any process that increases cell size. |
| positive regulation of interleukin-2 production | Any process that activates or increases the frequency, rate, or extent of interleukin-2 production. |
| positive regulation of JUN kinase activity | Any process that activates or increases the frequency, rate or extent of JUN kinase activity. |
| positive regulation of macroautophagy | Any process, such as recognition of nutrient depletion, that activates or increases the rate of macroautophagy to bring cytosolic macromolecules to the vacuole/lysosome for degradation. |
| positive regulation of non-canonical NF-kappaB signal transduction | Any process that activates or increases the frequency, rate or extent of NIK/NF-kappaB signaling. |
| positive regulation of T cell cytokine production | Any process that activates or increases the frequency, rate, or extent of T cell cytokine production. |
| positive regulation of vascular associated smooth muscle cell migration | Any process that activates or increases the frequency, rate or extent of vascular associated smooth muscle cell migration. |
| positive regulation of vascular associated smooth muscle cell proliferation | Any process that activates or increases the frequency, rate or extent of vascular smooth muscle cell proliferation. |
| stimulatory C-type lectin receptor signaling pathway | The series of molecular signals initiated by the binding of C-type lectin to its receptor on the surface of a target cell, and resulting in cellular activation. |
| stress-activated MAPK cascade | The series of molecular signals in which a stress-activated MAP kinase cascade relays a signal; MAP kinase cascades involve at least three protein kinase activities and culminate in the phosphorylation and activation of a MAP kinase. |
| T cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a T cell. |
| toll-like receptor 3 signaling pathway | The series of molecular signals initiated by a ligand binding to the endolysosomal toll-like receptor 3. |
| toll-like receptor 4 signaling pathway | The series of molecular signals initiated by a ligand binding to toll-like receptor 4. |
| transforming growth factor beta receptor signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a transforming growth factor beta receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| TRIF-dependent toll-like receptor signaling pathway | The series of molecular signals initiated by a ligand binding to a toll-like receptor where the TRIF adaptor mediates transduction of the signal. Toll-like receptors directly bind pattern motifs from a variety of microbial sources to initiate an innate immune response. |
37 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q4TVR5 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Bos taurus (Bovine) | PR |
| Q3SZJ2 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Bos taurus (Bovine) | PR |
| A2VDU3 | MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 | Bos taurus (Bovine) | SS |
| Q6XUX0 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Gallus gallus (Chicken) | PR |
| Q95UN8 | slpr | Mitogen-activated protein kinase kinase kinase | Drosophila melanogaster (Fruit fly) | EV |
| P83104 | Takl1 | Putative mitogen-activated protein kinase kinase kinase 7-like | Drosophila melanogaster (Fruit fly) | PR |
| O43353 | RIPK2 | Receptor-interacting serine/threonine-protein kinase 2 | Homo sapiens (Human) | PR |
| Q02779 | MAP3K10 | Mitogen-activated protein kinase kinase kinase 10 | Homo sapiens (Human) | SS |
| Q8NB16 | MLKL | Mixed lineage kinase domain-like protein | Homo sapiens (Human) | EV |
| Q16584 | MAP3K11 | Mitogen-activated protein kinase kinase kinase 11 | Homo sapiens (Human) | EV |
| P00540 | MOS | Proto-oncogene serine/threonine-protein kinase mos | Homo sapiens (Human) | PR |
| Q5TCX8 | MAP3K21 | Mitogen-activated protein kinase kinase kinase 21 | Homo sapiens (Human) | PR |
| Q6XUX3 | DSTYK | Dual serine/threonine and tyrosine protein kinase | Homo sapiens (Human) | PR |
| Q9NYL2 | MAP3K20 | Mitogen-activated protein kinase kinase kinase 20 | Homo sapiens (Human) | PR |
| Q38SD2 | LRRK1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Homo sapiens (Human) | EV |
| P80192 | MAP3K9 | Mitogen-activated protein kinase kinase kinase 9 | Homo sapiens (Human) | SS |
| Q80XI6 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Mus musculus (Mouse) | PR |
| Q8VDG6 | Map3k21 | Mitogen-activated protein kinase kinase kinase 21 | Mus musculus (Mouse) | PR |
| P58801 | Ripk2 | Receptor-interacting serine/threonine-protein kinase 2 | Mus musculus (Mouse) | PR |
| Q3U1V8 | Map3k9 | Mitogen-activated protein kinase kinase kinase 9 | Mus musculus (Mouse) | SS |
| Q9D2Y4 | Mlkl | Mixed lineage kinase domain-like protein | Mus musculus (Mouse) | SS |
| Q9ESL4 | Map3k20 | Mitogen-activated protein kinase kinase kinase 20 | Mus musculus (Mouse) | PR |
| P00536 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Mus musculus (Mouse) | PR |
| Q66L42 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Mus musculus (Mouse) | SS |
| Q62073 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Mus musculus (Mouse) | EV |
| Q66HA1 | Map3k11 | Mitogen-activated protein kinase kinase kinase 11 | Rattus norvegicus (Rat) | PR |
| D3ZG83 | Map3k10 | Mitogen-activated protein kinase kinase kinase 10 | Rattus norvegicus (Rat) | SS |
| P00539 | Mos | Proto-oncogene serine/threonine-protein kinase mos | Rattus norvegicus (Rat) | PR |
| P0C8E4 | Map3k7 | Mitogen-activated protein kinase kinase kinase 7 | Rattus norvegicus (Rat) | SS |
| Q9TZM3 | lrk-1 | Leucine-rich repeat serine/threonine-protein kinase 1 | Caenorhabditis elegans | SS |
| Q9FPR3 | EDR1 | Serine/threonine-protein kinase EDR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q05609 | CTR1 | Serine/threonine-protein kinase CTR1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q2MHE4 | HT1 | Serine/threonine/tyrosine-protein kinase HT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q67E00 | dstyk | Dual serine/threonine and tyrosine protein kinase | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSTASAASSS | SSSSAGEMIE | APSQVLNFEE | IDYKEIEVEE | VVGRGAFGVV | CKAKWRAKDV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AIKQIESESE | RKAFIVELRQ | LSRVNHPNIV | KLYGACLNPV | CLVMEYAEGG | SLYNVLHGAE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PLPYYTAAHA | MSWCLQCSQG | VAYLHSMQPK | ALIHRDLKPP | NLLLVAGGTV | LKICDFGTAC |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DIQTHMTNNK | GSAAWMAPEV | FEGSNYSEKC | DVFSWGIILW | EVITRRKPFD | EIGGPAFRIM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| WAVHNGTRPP | LIKNLPKPIE | SLMTRCWSKD | PSQRPSMEEI | VKIMTHLMRY | FPGADEPLQY |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PCQYSDEGQS | NSATSTGSFM | DIASTNTSNK | SDTNMEQVPA | TNDTIKRLES | KLLKNQAKQQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SESGRLSLGA | SRGSSVESLP | PTSEGKRMSA | DMSEIEARIA | ATTAYSKPKR | GHRKTASFGN |
| 430 | 440 | 450 | 460 | 470 | 480 |
| ILDVPEIVIS | GNGQPRRRSI | QDLTVTGTEP | GQVSSRSSSP | SVRMITTSGP | TSEKPTRSHP |
| 490 | 500 | 510 | 520 | 530 | 540 |
| WTPDDSTDTN | GSDNSIPMAY | LTLDHQLQPL | APCPNSKESM | AVFEQHCKMA | QEYMKVQTEI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ALLLQRKQEL | VAELDQDEKD | QQNTSRLVQE | HKKLLDENKS | LSTYYQQCKK | QLEVIRSQQQ |
| KRQGTS |