O35963
Gene name |
Rab33b |
Protein name |
Ras-related protein Rab-33B |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:19338 |
EC number |
|
Protein Class |
|
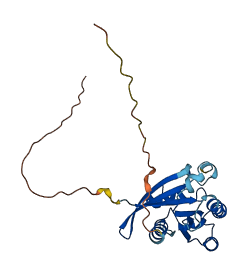
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for O35963
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1Z06 | X-ray | 181 A | A | 30-202 | PDB |
| 2G77 | X-ray | 226 A | B | 14-202 | PDB |
| 6SUR | X-ray | 347 A | A/B/C/D/E/F | 30-202 | PDB |
| AF-O35963-F1 | Predicted | AlphaFoldDB |
11 variants for O35963
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388612571 | 9 | L>Q | No | EVA | |
| rs3393039623 | 65 | T>A | No | EVA | |
| rs3393039625 | 66 | I>V | No | EVA | |
| rs3393039627 | 68 | V>E | No | EVA | |
| rs3388612589 | 94 | R>G | No | EVA | |
| rs3388633609 | 110 | V>I | No | EVA | |
| rs3388625271 | 169 | T>I | No | EVA | |
| rs3388617838 | 172 | M>I | No | EVA | |
| rs3388612578 | 179 | A>V | No | EVA | |
| rs3388630159 | 185 | N>T | No | EVA | |
| rs3388625684 | 186 | D>H | No | EVA |
No associated diseases with O35963
1 regional properties for O35963
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 33 - 181 | IPR005225 |
Functions
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi lumen | The volume enclosed by the membranes of any cisterna or subcompartment of the Golgi apparatus, including the cis- and trans-Golgi networks. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| presynapse | The part of a synapse that is part of the presynaptic cell. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
10 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome assembly | The formation of a double membrane-bounded structure, the autophagosome, that occurs when a specialized membrane sac, called the isolation membrane, starts to enclose a portion of the cytoplasm. |
| intra-Golgi vesicle-mediated transport | The directed movement of substances within the Golgi, mediated by small transport vesicles. These either fuse with the cis-Golgi or with each other to form the membrane stacks known as the cis-Golgi reticulum (network). |
| negative regulation of constitutive secretory pathway | Any process that stops, prevents or reduces the frequency, rate or extent of constitutive secretory pathway. |
| protein localization to Golgi apparatus | A process in which a protein is transported to, or maintained in, a location within the Golgi apparatus. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| Rab protein signal transduction | The series of molecular signals within the cell that are mediated by a member of the Rab family of proteins switching to a GTP-bound active state. |
| regulation of exocytosis | Any process that modulates the frequency, rate or extent of exocytosis. |
| regulation of Golgi organization | Any process that modulates the frequency, rate or extent of Golgi organization. |
| regulation of retrograde vesicle-mediated transport, Golgi to ER | Any process that modulates the frequency, rate or extent of retrograde vesicle-mediated transport, Golgi to ER. |
| skeletal system morphogenesis | The process in which the anatomical structures of the skeleton are generated and organized. |
15 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3ZC27 | RAB19 | Ras-related protein Rab-19 | Bos taurus (Bovine) | PR |
| Q5ZHV1 | RAB33B | Ras-related protein Rab-33B | Gallus gallus (Chicken) | PR |
| A4D1S5 | RAB19 | Ras-related protein Rab-19 | Homo sapiens (Human) | PR |
| Q9H082 | RAB33B | Ras-related protein Rab-33B | Homo sapiens (Human) | PR |
| P35294 | Rab19 | Ras-related protein Rab-19 | Mus musculus (Mouse) | PR |
| Q9JKM7 | Rab37 | Ras-related protein Rab-37 | Mus musculus (Mouse) | PR |
| Q5M7U5 | Rab19 | Ras-related protein Rab-19 | Rattus norvegicus (Rat) | PR |
| P25766 | RGP1 | Ras-related protein RGP1 | Oryza sativa subsp japonica (Rice) | PR |
| Q20365 | rab-33 | Ras-related protein Rab-33 | Caenorhabditis elegans | PR |
| Q9LNK1 | RABA3 | Ras-related protein RABA3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O04486 | RABA2A | Ras-related protein RABA2a | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LNW1 | RABA2B | Ras-related protein RABA2b | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LH50 | RABA4D | Ras-related protein RABA4d | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FE79 | RABA4C | Ras-related protein RABA4c | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SJZ5 | RABA4E | Putative Ras-related protein RABA4e | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTSEMESSLE | VSFSSSCAVS | GASGCLPPAR | SRIFKIIVIG | DSNVGKTCLT | YRFCAGRFPD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| RTEATIGVDF | RERAVDIDGE | RIKIQLWDTA | GQERFRKSMV | QHYYRNVHAV | VFVYDMTNMA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SFHSLPAWIE | ECKQHLLAND | IPRILVGNKC | DLRSAIQVPT | DLAQKFADTH | SMPLFETSAK |
| 190 | 200 | 210 | 220 | ||
| NPNDNDHVEA | IFMTLAHKLK | SHKPLMLSQL | PDNRISLKPE | TKPAVTCWC |