O35942
Gene name |
Nek2 |
Protein name |
Serine/threonine-protein kinase Nek2 |
Names |
Never in mitosis A-related kinase 2, NimA-related protein kinase 2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:18005 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
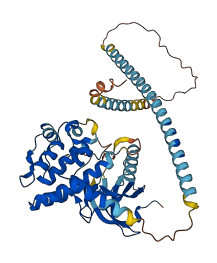
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
158-181 (Activation loop from InterPro)
Target domain |
8-271 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for O35942
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O35942-F1 | Predicted | AlphaFoldDB |
23 variants for O35942
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388525961 | 37 | K>E | No | EVA | |
| rs220933299 | 47 | V>M | No | EVA | |
| rs3391190533 | 69 | S>N | No | EVA | |
| rs31377073 | 69 | S>R | No | EVA | |
| rs3388521327 | 94 | L>Q | No | EVA | |
| rs49224556 | 95 | A>G | No | EVA | |
| rs51716376 | 186 | E>D | No | EVA | |
| rs255625300 | 188 | M>I | No | EVA | |
| rs3391163263 | 194 | N>K | No | EVA | |
| rs3391158821 | 250 | R>W | No | EVA | |
| rs31963153 | 274 | L>M | No | EVA | |
| rs3388526972 | 297 | P>S | No | EVA | |
| rs3391188004 | 305 | E>* | No | EVA | |
| rs48235999 | 311 | R>S | No | EVA | |
| rs3388525926 | 322 | A>S | No | EVA | |
| rs3388526308 | 327 | L>M | No | EVA | |
| rs3388521369 | 340 | L>H | No | EVA | |
| rs3388522690 | 347 | R>I | No | EVA | |
| rs3388524594 | 353 | K>Q | No | EVA | |
| rs223367009 | 356 | S>I | No | EVA | |
| rs223367009 | 356 | S>N | No | EVA | |
| rs3391124689 | 362 | R>W | No | EVA | |
| rs3388521360 | 398 | N>K | No | EVA |
No associated diseases with O35942
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| condensed nuclear chromosome | A highly compacted molecule of DNA and associated proteins resulting in a cytologically distinct nuclear chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| kinetochore | A multisubunit complex that is located at the centromeric region of DNA and provides an attachment point for the spindle microtubules. |
| microtubule | Any of the long, generally straight, hollow tubes of internal diameter 12-15 nm and external diameter 24 nm found in a wide variety of eukaryotic cells; each consists (usually) of 13 protofilaments of polymeric tubulin, staggered in such a manner that the tubulin monomers are arranged in a helical pattern on the microtubular surface, and with the alpha/beta axes of the tubulin subunits parallel to the long axis of the tubule; exist in equilibrium with pool of tubulin monomers and can be rapidly assembled or disassembled in response to physiological stimuli; concerned with force generation, e.g. in the spindle. |
| midbody | A thin cytoplasmic bridge formed between daughter cells at the end of cytokinesis. The midbody forms where the contractile ring constricts, and may persist for some time before finally breaking to complete cytokinesis. |
| nucleolus | A small, dense body one or more of which are present in the nucleus of eukaryotic cells. It is rich in RNA and protein, is not bounded by a limiting membrane, and is not seen during mitosis. Its prime function is the transcription of the nucleolar DNA into 45S ribosomal-precursor RNA, the processing of this RNA into 5.8S, 18S, and 28S components of ribosomal RNA, and the association of these components with 5S RNA and proteins synthesized outside the nucleolus. This association results in the formation of ribonucleoprotein precursors; these pass into the cytoplasm and mature into the 40S and 60S subunits of the ribosome. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| spindle pole | Either of the ends of a spindle, where spindle microtubules are organized; usually contains a microtubule organizing center and accessory molecules, spindle microtubules and astral microtubules. |
6 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| metal ion binding | Binding to a metal ion. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein phosphatase binding | Binding to a protein phosphatase. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| blastocyst development | The process whose specific outcome is the progression of the blastocyst over time, from its formation to the mature structure. The mammalian blastocyst is a hollow ball of cells containing two cell types, the inner cell mass and the trophectoderm. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| centrosome separation | The process in which duplicated centrosome components move away from each other. The centriole pair within each centrosome becomes part of a separate microtubule organizing center that nucleates a radial array of microtubules called an aster. The two asters move to opposite sides of the nucleus to form the two poles of the mitotic spindle. |
| chromosome segregation | The process in which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets. In eukaryotes, chromosome segregation begins with the condensation of chromosomes, includes chromosome separation, and ends when chromosomes have completed movement to the spindle poles. |
| meiotic cell cycle | Progression through the phases of the meiotic cell cycle, in which canonically a cell replicates to produce four offspring with half the chromosomal content of the progenitor cell via two nuclear divisions. |
| mitotic sister chromatid segregation | The cell cycle process in which replicated homologous chromosomes are organized and then physically separated and apportioned to two sets during the mitotic cell cycle. Each replicated chromosome, composed of two sister chromatids, aligns at the cell equator, paired with its homologous partner. One homolog of each morphologic type goes into each of the resulting chromosome sets. |
| mitotic spindle assembly | Mitotic bipolar spindle assembly begins with spindle microtubule nucleation from the separated spindle pole body, includes spindle elongation during prometaphase, and is complete when all kinetochores are stably attached the spindle, and the spindle assembly checkpoint is satisfied. |
| negative regulation of centriole-centriole cohesion | Any process that stops, prevents or reduces the frequency, rate or extent of centriole-centriole cohesion. |
| negative regulation of DNA binding | Any process that stops or reduces the frequency, rate or extent of DNA binding. DNA binding is any process in which a gene product interacts selectively with DNA (deoxyribonucleic acid). |
| positive regulation of telomerase activity | Any process that activates or increases the frequency, rate or extent of telomerase activity, the catalysis of the reaction: deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
| positive regulation of telomere capping | Any process that activates or increases the frequency, rate or extent of telomere capping. |
| positive regulation of telomere maintenance via telomerase | Any process that activates or increases the frequency, rate or extent of the addition of telomeric repeats by telomerase. |
| protein autophosphorylation | The phosphorylation by a protein of one or more of its own amino acid residues (cis-autophosphorylation), or residues on an identical protein (trans-autophosphorylation). |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of attachment of spindle microtubules to kinetochore | Any process that modulates the frequency, rate or extent of the attachment of spindle microtubules to the kinetochore. |
| regulation of mitotic centrosome separation | Any process that modulates the frequency, rate or extent of the separation of duplicated centrosome components at the beginning of mitosis. |
9 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P22209 | KIN3 | Serine/threonine-protein kinase KIN3 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P21359 | NF1 | Neurofibromin | Homo sapiens (Human) | PR |
| P51955 | NEK2 | Serine/threonine-protein kinase Nek2 | Homo sapiens (Human) | EV |
| Q04690 | Nf1 | Neurofibromin | Mus musculus (Mouse) | PR |
| Q6ZEZ5 | NEK3 | Serine/threonine-protein kinase Nek3 | Oryza sativa subsp japonica (Rice) | PR |
| Q10GB1 | NEK1 | Serine/threonine-protein kinase Nek1 | Oryza sativa subsp japonica (Rice) | PR |
| Q9SLI2 | NEK1 | Serine/threonine-protein kinase Nek1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RXT4 | NEK4 | Serine/threonine-protein kinase Nek4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LT35 | NEK6 | Serine/threonine-protein kinase Nek6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MPSRVEDYEV | LHSIGTGSYG | RCQKIRRKSD | GKILVWKELD | YGSMTEVEKQ | MLVSEVNLLR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ELKHPNIVSY | YDRIIDRTNT | TLYIVMEYCE | GGDLASVISK | GTKDRQYLEE | EFVLRVMTQL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| TLALKECHRR | SDGGHTVLHR | DLKPANVFLD | SKHNVKLGDF | GLARILNHDT | SFAKTFVGTP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YYMSPEQMSC | LSYNEKSDIW | SLGCLLYELC | ALMPPFTAFN | QKELAGKIRE | GRFRRIPYRY |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SDGLNDLITR | MLNLKDYHRP | SVEEILESPL | IADLVAEEQR | RNLERRGRRS | GEPSKLPDSS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PVLSELKLKE | RQLQDREQAL | RAREDILEQK | ERELCIRERL | AEDKLARAES | LMKNYSLLKE |
| 370 | 380 | 390 | 400 | 410 | 420 |
| HRLLCLAGGP | ELDLPSSAMK | KKVHFHGESK | ENTARSENSE | SYLAKSKCRD | LKKRLHAAQL |
| 430 | 440 | ||||
| RAQALADIEK | NYQLKSRQIL | GMR |