O35618
Gene name |
Mdm4 (Mdmx) |
Protein name |
Protein Mdm4 |
Names |
Double minute 4 protein, Mdm2-like p53-binding protein, Protein Mdmx, p53-binding protein Mdm4 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:17248 |
EC number |
|
Protein Class |
|
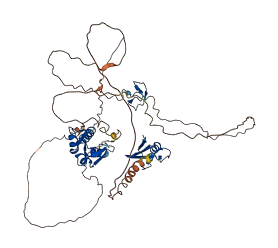
Descriptions
MDM4 is a homologous protein that binds to p53 and regulates its activity. Intrinsically disordered region constitute an inhibitory module. It competes with the p53TAD for binding to the NTD of MDM4, thus lowering the affinity of MDMX for p53. Efficient binding of MDM4 to p53 requires a mechanism to relieve the allosteric self-inhibition.
Autoinhibitory domains (AIDs)
Target domain |
25-108 (SWIB/MDM2 domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for O35618
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O35618-F1 | Predicted | AlphaFoldDB |
7 variants for O35618
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs6192152 | 112 | I>N | No | Ensembl | |
| rs253523478 | 210 | C>Y | No | Ensembl | |
| rs582516334 | 274 | T>M | No | Ensembl | |
| rs245555288 | 401 | E>G | No | Ensembl | |
| rs243387396 | 409 | A>E | No | Ensembl | |
| rs218592325 | 411 | E>G | No | Ensembl | |
| rs223796411 | 475 | A>V | No | Ensembl |
No associated diseases with O35618
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| metal ion binding | Binding to a metal ion. |
| p53 binding | Binding to one of the p53 family of proteins. |
14 GO annotations of biological process
| Name | Definition |
|---|---|
| atrial septum development | The progression of the atrial septum over time, from its initial formation to the mature structure. |
| atrioventricular valve morphogenesis | The process in which the structure of the atrioventricular valve is generated and organized. |
| DNA damage response, signal transduction by p53 class mediator | A cascade of processes induced by the cell cycle regulator phosphoprotein p53, or an equivalent protein, in response to the detection of DNA damage. |
| endocardial cushion morphogenesis | The process in which the anatomical structure of the endocardial cushion is generated and organized. The endocardial cushion is a specialized region of mesenchymal cells that will give rise to the heart septa and valves. |
| heart valve development | The progression of a heart valve over time, from its formation to the mature structure. A heart valve is a structure that restricts the flow of blood to different regions of the heart and forms from an endocardial cushion. |
| negative regulation of intrinsic apoptotic signaling pathway by p53 class mediator | Any process that stops, prevents or reduces the frequency, rate or extent of intrinsic apoptotic signaling pathway by p53 class mediator. |
| negative regulation of protein catabolic process | Any process that stops, prevents or reduces the frequency, rate or extent of protein catabolic process. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| protein-containing complex assembly | The aggregation, arrangement and bonding together of a set of macromolecules to form a protein-containing complex. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of heart rate | Any process that modulates the frequency or rate of heart contraction. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| ventricular septum development | The progression of the ventricular septum over time from its formation to the mature structure. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q2HJ21 | MDM4 | Protein Mdm4 | Bos taurus (Bovine) | SS |
| E1C4B0 | MDM4 | Double minute 4 protein | Gallus gallus (Chicken) | SS |
| O15151 | MDM4 | Protein Mdm4 | Homo sapiens (Human) | EV |
| Q8VCM5 | Mul1 | Mitochondrial ubiquitin ligase activator of NFKB 1 | Mus musculus (Mouse) | PR |
| Q5XIN1 | Mdm4 | Protein Mdm4 | Rattus norvegicus (Rat) | SS |
| Q7ZUW7 | mdm4 | Protein Mdm4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTSHSTSAQC | SASDSACRIS | SEQISQVRPK | LQLLKILHAA | GAQGEVFTMK | EVMHYLGQYI |
| 70 | 80 | 90 | 100 | 110 | 120 |
| MVKQLYDQQE | QHMVYCGGDL | LGDLLGCQSF | SVKDPSPLYD | MLRKNLVTSA | SINTDAAQTL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ALAQDHTMDF | PSQDRLKHGA | TEYSNPRKRT | EEEDTHTLPT | SRHKCRDSRA | DEDLIEHLSQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| DETSRLDLDF | EEWDVAGLPW | WFLGNLRNNC | IPKSNGSTDL | QTNQDIGTAI | VSDTTDDLWF |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LNETVSEQLG | VGIKVEAANS | EQTSEVGKTS | NKKTVEVGKD | DDLEDSRSLS | DDTDVELTSE |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DEWQCTECKK | FNSPSKRYCF | RCWALRKDWY | SDCSKLTHSL | STSNITAIPE | KKDNEGIDVP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| DCRRTISAPV | VRPKDGYLKE | EKPRFDPCNS | VGFLDLAHSS | ESQEIISSAR | EQTDIFSEQK |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AETESMEDFQ | NVLKPCSLCE | KRPRDGNIIH | GKTSHLTTCF | HCARRLKKSG | ASCPACKKEI |
| QLVIKVFIA |