O31516
Gene name |
yesM (BSU06950) |
Protein name |
Sensor histidine kinase YesM |
Names |
|
Species |
Bacillus subtilis (strain 168) |
KEGG Pathway |
bsu:BSU06950 |
EC number |
2.7.13.3: Protein-histidine kinases |
Protein Class |
|
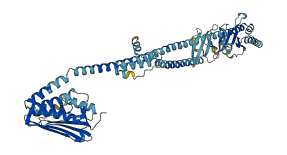
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for O31516
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O31516-F1 | Predicted | AlphaFoldDB |
No variants for O31516
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for O31516 | |||||
No associated diseases with O31516
1 regional properties for O31516
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 3 - 164 | IPR005225 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.13.3 | Protein-histidine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| phosphorelay sensor kinase activity | Catalysis of the phosphorylation of a histidine residue in response to detection of an extracellular signal such as a chemical ligand or change in environment, to initiate a change in cell state or activity. The two-component sensor is a histidine kinase that autophosphorylates a histidine residue in its active site. The phosphate is then transferred to an aspartate residue in a downstream response regulator, to trigger a response. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKKRVAGWYR | RMKIKDKLFV | FLSLIMAVSF | LFVYSGVQYA | FHVYDEQIYR | KSSEVLRMSS |
| 70 | 80 | 90 | 100 | 110 | 120 |
| ERIEDELKKI | EDVSYEIITD | EQIQRILSMQ | NRDDTYDQYQ | MKQELWDQLA | GYASDEKYID |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SIHVIDARGS | EYSAGSSSSD | LLQQEQEEVF | KRAKAKSGRN | LWMTLGGSDP | VLISARQIRS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| YHQLSLNGLG | MVLIQVNVKQ | MIRDVPKDWG | DSVGDIMIAD | QGGNLVYTAH | ASAHVPEAAK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ETLKHPGYDL | IKKNGKRYFI | SYLQSSYQNW | SYYNVIPFDQ | MFAKISFMKT | VIGTCFLLFF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| CVVLLFGRKI | ANSITEPIEQ | LVTAMKSVQH | SGIEAGVSLS | LPEHTQDEAG | MLNRHFTVMM |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KRINELMEEN | VEKQLIIKET | ELKALQAQIN | PHFLYNTLES | INWLAKANQQ | KQISKMVESL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GFLLRNSIHM | KKDIVTIQEE | ADIVRHYMTI | QRFRFEERLK | FTLDIDDEVK | HCLIPKLTLQ |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PLAENAIQYA | LEPFTRPCAI | RIQAKKAKGC | VCITVEDNGP | GMDGRILEST | GGRGIGLWNI |
| 550 | 560 | 570 | |||
| RERISLTFGE | PYGLRIHSEH | EKGTRIVITI | PCRNEVV |