O23447
Gene name |
PME43 (ARATH43, At4g15980, dl4030c, FCAALL.248) |
Protein name |
Putative pectinesterase/pectinesterase inhibitor 43 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT4G15980 |
EC number |
3.1.1.11: Carboxylic ester hydrolases |
Protein Class |
|
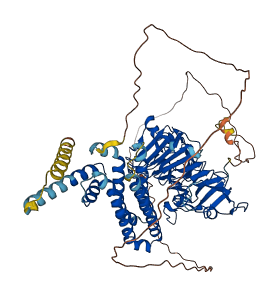
Descriptions
(Annotation from UniProt)
The PMEI region may act as an autoinhibitory domain.
Autoinhibitory domains (AIDs)
Target domain |
391-688 (Pectinesterase domain) |
Relief mechanism |
|
Assay |
|
Accessory elements
No accessory elements
References
Autoinhibited structure

Activated structure

1 structures for O23447
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O23447-F1 | Predicted | AlphaFoldDB |
174 variants for O23447
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH02848418 | 2 | N>K | No | 1000Genomes | |
| ENSVATH06683405 | 4 | Y>S | No | 1000Genomes | |
| tmp_4_9059966_C_T,G | 6 | L>F | No | 1000Genomes | |
| tmp_4_9059968_A_C | 6 | L>V | No | 1000Genomes | |
| tmp_4_9059955_G_A | 10 | T>M | No | 1000Genomes | |
| ENSVATH14210937 | 11 | A>V | No | 1000Genomes | |
| ENSVATH14210936 | 15 | A>E | No | 1000Genomes | |
| ENSVATH14210936 | 15 | A>G | No | 1000Genomes | |
| tmp_4_9059919_G_T | 22 | A>D | No | 1000Genomes | |
| tmp_4_9059915_G_T | 23 | N>K | No | 1000Genomes | |
| tmp_4_9059907_T_C | 26 | N>S | No | 1000Genomes | |
| tmp_4_9059888_C_T | 32 | M>I | No | 1000Genomes | |
| ENSVATH02848417 | 38 | E>K | No | 1000Genomes | |
| ENSVATH06683402 | 38 | E>V | No | 1000Genomes | |
| tmp_4_9059851_T_A | 45 | K>* | No | 1000Genomes | |
| ENSVATH06683401 | 45 | K>T | No | 1000Genomes | |
| ENSVATH11820897 | 54 | S>F | No | 1000Genomes | |
| tmp_4_9059813_G_T | 57 | Y>* | No | 1000Genomes | |
| ENSVATH06683399 | 62 | T>I | No | 1000Genomes | |
| ENSVATH06683398 | 66 | A>T | No | 1000Genomes | |
| ENSVATH02848416 | 67 | T>A | No | 1000Genomes | |
| tmp_4_9059755_T_A | 77 | I>L | No | 1000Genomes | |
| tmp_4_9059737_G_T | 83 | L>I | No | 1000Genomes | |
| tmp_4_9059727_A_G | 86 | I>T | No | 1000Genomes | |
| tmp_4_9059728_T_C | 86 | I>V | No | 1000Genomes | |
| tmp_4_9059718_C_T | 89 | R>Q | No | 1000Genomes | |
| ENSVATH06683395 | 91 | G>R | No | 1000Genomes | |
| ENSVATH06683394 | 96 | M>I | No | 1000Genomes | |
| ENSVATH06683393 | 100 | K>N | No | 1000Genomes | |
| ENSVATH11820895 | 108 | H>Y | No | 1000Genomes | |
| ENSVATH06683390 | 112 | A>S | No | 1000Genomes | |
| ENSVATH06683390 | 112 | A>T | No | 1000Genomes | |
| ENSVATH06683389 | 112 | A>V | No | 1000Genomes | |
| ENSVATH06683388 | 115 | T>I | No | 1000Genomes | |
| tmp_4_9059620_C_T | 122 | D>N | No | 1000Genomes | |
| tmp_4_9059611_C_G | 125 | D>H | No | 1000Genomes | |
| ENSVATH06683386 | 137 | F>Y | No | 1000Genomes | |
| ENSVATH02848415 | 140 | T>S | No | 1000Genomes | |
| tmp_4_9059562_C_T | 141 | R>K | No | 1000Genomes | |
| tmp_4_9059528_C_G | 152 | W>C | No | 1000Genomes | |
| tmp_4_9059479_C_G | 169 | G>R | No | 1000Genomes | |
| ENSVATH00510189 | 177 | M>I | No | 1000Genomes | |
| tmp_4_9059433_T_C | 184 | K>R | No | 1000Genomes | |
| ENSVATH02848414 | 185 | G>A | No | 1000Genomes | |
| ENSVATH06683382 | 194 | A>G | No | 1000Genomes | |
| tmp_4_9059398_C_T | 196 | A>T | No | 1000Genomes | |
| tmp_4_9059372_C_G,T | 204 | K>N | No | 1000Genomes | |
| tmp_4_9059347_G_A | 213 | L>F | No | 1000Genomes | |
| ENSVATH00510185 | 216 | R>K | No | 1000Genomes | |
| tmp_4_9059193_A_T | 223 | L>* | No | 1000Genomes | |
| ENSVATH14210883 | 224 | G>R | No | 1000Genomes | |
| tmp_4_9059184_C_G | 226 | G>A | No | 1000Genomes | |
| ENSVATH06683375 | 227 | S>T | No | 1000Genomes | |
| ENSVATH00510184 | 231 | E>Q | No | 1000Genomes | |
| tmp_4_9059166_G_A | 232 | S>L | No | 1000Genomes | |
| tmp_4_9059157_G_A | 235 | S>F | No | 1000Genomes | |
| ENSVATH02848405 | 239 | S>Y | No | 1000Genomes | |
| ENSVATH06683374 | 246 | S>F | No | 1000Genomes | |
| tmp_4_9059112_G_C | 250 | P>R | No | 1000Genomes | |
| ENSVATH00510181 | 251 | S>I | No | 1000Genomes | |
| ENSVATH06683372 | 254 | D>E | No | 1000Genomes | |
| ENSVATH11820797 | 256 | S>L | No | 1000Genomes | |
| tmp_4_9059080_C_A | 261 | V>L | No | 1000Genomes | |
| tmp_4_9059070_G_C | 264 | S>* | No | 1000Genomes | |
| tmp_4_9059068_A_C | 265 | S>A | No | 1000Genomes | |
| tmp_4_9059067_G_A | 265 | S>L | No | 1000Genomes | |
| ENSVATH06683371 | 266 | E>G | No | 1000Genomes | |
| tmp_4_9059059_G_T | 268 | Q>K | No | 1000Genomes | |
| tmp_4_9059058_T_A | 268 | Q>L | No | 1000Genomes | |
| ENSVATH00510179 | 269 | S>P | No | 1000Genomes | |
| ENSVATH06683369 | 273 | S>L | No | 1000Genomes | |
| ENSVATH06683370 | 273 | S>P | No | 1000Genomes | |
| ENSVATH06683368 | 274 | N>D | No | 1000Genomes | |
| ENSVATH06683367 | 274 | N>S | No | 1000Genomes | |
| ENSVATH06683366 | 277 | P>Q | No | 1000Genomes | |
| tmp_4_9059032_G_T | 277 | P>T | No | 1000Genomes | |
| tmp_4_9058998_G_T | 288 | S>Y | No | 1000Genomes | |
| tmp_4_9058995_G_A | 289 | S>F | No | 1000Genomes | |
| ENSVATH06683365 | 290 | E>Q | No | 1000Genomes | |
| ENSVATH06683364 | 291 | D>N | No | 1000Genomes | |
| tmp_4_9058983_G_C | 293 | P>R | No | 1000Genomes | |
| tmp_4_9058979_T_G | 294 | K>N | No | 1000Genomes | |
| ENSVATH06683361 | 295 | K>N | No | 1000Genomes | |
| ENSVATH06683362 | 295 | K>R | No | 1000Genomes | |
| tmp_4_9058975_A_T | 296 | S>T | No | 1000Genomes | |
| ENSVATH00510177 | 298 | F>S | No | 1000Genomes | |
| tmp_4_9058965_G_A | 299 | S>L | No | 1000Genomes | |
| ENSVATH00510176 | 300 | G>E | No | 1000Genomes | |
| tmp_4_9058954_G_T | 303 | P>T | No | 1000Genomes | |
| ENSVATH06683359 | 305 | D>N | No | 1000Genomes | |
| tmp_4_9058919_T_A | 314 | K>N | No | 1000Genomes | |
| ENSVATH00510175 | 315 | S>P | No | 1000Genomes | |
| ENSVATH06683358 | 316 | T>S | No | 1000Genomes | |
| ENSVATH11820783 | 318 | S>C | No | 1000Genomes | |
| ENSVATH11820784 | 318 | S>T | No | 1000Genomes | |
| ENSVATH00510174 | 320 | N>I | No | 1000Genomes | |
| tmp_4_9058899_T_C | 321 | Q>R | No | 1000Genomes | |
| ENSVATH06683355 | 322 | P>Q | No | 1000Genomes | |
| ENSVATH06683354 | 323 | L>F | No | 1000Genomes | |
| ENSVATH06683353 | 324 | D>N | No | 1000Genomes | |
| tmp_4_9058884_G_A | 326 | S>F | No | 1000Genomes | |
| ENSVATH11820781 | 327 | E>D | No | 1000Genomes | |
| ENSVATH11820780 | 328 | N>K | No | 1000Genomes | |
| ENSVATH11820779 | 329 | P>L | No | 1000Genomes | |
| ENSVATH11820778 | 330 | P>L | No | 1000Genomes | |
| tmp_4_9058864_A_T | 333 | S>T | No | 1000Genomes | |
| tmp_4_9058857_G_A | 335 | S>F | No | 1000Genomes | |
| tmp_4_9058831_G_C | 344 | L>V | No | 1000Genomes | |
| ENSVATH11820760 | 345 | R>C | No | 1000Genomes | |
| tmp_4_9058827_C_T | 345 | R>H | No | 1000Genomes | |
| tmp_4_9058820_C_G | 347 | L>F | No | 1000Genomes | |
| ENSVATH02848404 | 351 | N>K | No | 1000Genomes | |
| ENSVATH02848404 | 351 | N>K | No | 1000Genomes | |
| tmp_4_9058807_T_G | 352 | K>Q | No | 1000Genomes | |
| tmp_4_9058802_C_A | 353 | L>F | No | 1000Genomes | |
| tmp_4_9058800_T_C | 354 | D>G | No | 1000Genomes | |
| ENSVATH06683351 | 363 | E>K | No | 1000Genomes | |
| ENSVATH06683351 | 363 | E>Q | No | 1000Genomes | |
| ENSVATH11820759 | 364 | E>A | No | 1000Genomes | |
| ENSVATH11820758 | 373 | P>S | No | 1000Genomes | |
| tmp_4_9058734_C_T | 376 | R>Q | No | 1000Genomes | |
| tmp_4_9058731_C_T | 377 | R>K | No | 1000Genomes | |
| ENSVATH00510172 | 384 | R>K | No | 1000Genomes | |
| ENSVATH00510172 | 384 | R>T | No | 1000Genomes | |
| ENSVATH11820757 | 387 | G>E | No | 1000Genomes | |
| tmp_4_9058695_T_C | 389 | K>R | No | 1000Genomes | |
| tmp_4_9058692_G_C | 390 | A>G | No | 1000Genomes | |
| ENSVATH06683350 | 395 | A>T | No | 1000Genomes | |
| ENSVATH06683349 | 398 | G>R | No | 1000Genomes | |
| tmp_4_9058662_C_T | 400 | G>D | No | 1000Genomes | |
| tmp_4_9058644_G_A | 406 | A>V | No | 1000Genomes | |
| tmp_4_9058642_G_A | 407 | Q>* | No | 1000Genomes | |
| tmp_4_9058611_G_A | 417 | T>I | No | 1000Genomes | |
| tmp_4_9058604_C_G | 419 | K>N | No | 1000Genomes | |
| tmp_4_9058591_T_G | 424 | I>L | No | 1000Genomes | |
| tmp_4_9058561_C_G | 434 | E>Q | No | 1000Genomes | |
| tmp_4_9058551_T_C | 437 | K>R | No | 1000Genomes | |
| ENSVATH06683348 | 440 | L>V | No | 1000Genomes | |
| tmp_4_9058512_G_T | 450 | T>K | No | 1000Genomes | |
| tmp_4_9058504_C_T | 453 | V>I | No | 1000Genomes | |
| ENSVATH11820755 | 466 | G>S | No | 1000Genomes | |
| tmp_4_9058458_T_A | 468 | Y>F | No | 1000Genomes | |
| tmp_4_9058456_G_A,C | 469 | R>C | No | 1000Genomes | |
| tmp_4_9058456_G_A,C | 469 | R>G | No | 1000Genomes | |
| ENSVATH00510168 | 473 | V>F | No | 1000Genomes | |
| tmp_4_9058228_A_G | 479 | Y>H | No | 1000Genomes | |
| ENSVATH11820728 | 481 | M>I | No | 1000Genomes | |
| tmp_4_9058208_T_C | 485 | I>M | No | 1000Genomes | |
| tmp_4_9058147_C_A | 506 | D>Y | No | 1000Genomes | |
| tmp_4_9058143_A_G | 507 | F>S | No | 1000Genomes | |
| ENSVATH11820726 | 527 | H>N | No | 1000Genomes | |
| ENSVATH14210878 | 530 | F>L | No | 1000Genomes | |
| tmp_4_9058056_A_T | 536 | V>D | No | 1000Genomes | |
| ENSVATH14210877 | 538 | G>S | No | 1000Genomes | |
| ENSVATH06683341 | 553 | N>K | No | 1000Genomes | |
| ENSVATH00510162 | 557 | V>I | No | 1000Genomes | |
| tmp_4_9057985_G_A | 560 | R>C | No | 1000Genomes | |
| ENSVATH06683340 | 564 | H>N | No | 1000Genomes | |
| tmp_4_9057971_G_C,A | 564 | H>Q | No | 1000Genomes | |
| ENSVATH06683340 | 564 | H>Y | No | 1000Genomes | |
| ENSVATH00510161 | 566 | Q>R | No | 1000Genomes | |
| ENSVATH06683338 | 577 | R>W | No | 1000Genomes | |
| ENSVATH11820674 | 591 | T>I | No | 1000Genomes | |
| tmp_4_9057889_C_T | 592 | G>S | No | 1000Genomes | |
| tmp_4_9057880_A_C | 595 | S>A | No | 1000Genomes | |
| ENSVATH11820671 | 631 | E>K | No | 1000Genomes | |
| tmp_4_9057671_A_G | 634 | L>S | No | 1000Genomes | |
| tmp_4_9057609_G_C | 655 | R>G | No | 1000Genomes | |
| tmp_4_9057590_C_A | 661 | R>L | No | 1000Genomes | |
| tmp_4_9057575_C_A | 666 | R>I | No | 1000Genomes | |
| ENSVATH06683330 | 669 | K>E | No | 1000Genomes | |
| tmp_4_9057542_C_G | 677 | R>T | No | 1000Genomes | |
| ENSVATH00510157 | 689 | T>I | No | 1000Genomes | |
| ENSVATH06683328 | 693 | Q>H | No | 1000Genomes |
No associated diseases with O23447
10 regional properties for O23447
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 609 - 737 | IPR000008 |
| domain | Phosphatidylinositol-specific phospholipase C, X domain | 296 - 441 | IPR000909 |
| domain | Phospholipase C, phosphatidylinositol-specific, Y domain | 492 - 609 | IPR001711 |
| domain | Pleckstrin homology domain | 19 - 132 | IPR001849 |
| domain | EF-hand domain | 140 - 175 | IPR002048 |
| domain | Phosphoinositide-specific phospholipase C, EF-hand-like domain | 204 - 288 | IPR015359 |
| binding_site | EF-Hand 1, calcium-binding site | 153 - 165 | IPR018247-1 |
| binding_site | EF-Hand 1, calcium-binding site | 189 - 201 | IPR018247-2 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1, catalytic domain | 295 - 596 | IPR028391 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1, EF-hand domain | 144 - 283 | IPR046975 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.1.11 | Carboxylic ester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| apoplast | The cell membranes and intracellular regions in a plant are connected through plasmodesmata, and plants may be described as having two major compartments: the living symplast and the non-living apoplast. The apoplast is external to the plasma membrane and includes cell walls, intercellular spaces and the lumen of dead structures such as xylem vessels. Water and solutes pass freely through it. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| aspartyl esterase activity | Catalysis of the hydrolysis of an ester bond by a mechanism involving a catalytically active aspartic acid residue. |
| pectinesterase activity | Catalysis of the reaction: pectin + n H2O = n methanol + pectate. |
| pectinesterase inhibitor activity | Binds to and stops, prevents or reduces the activity of pectinesterase. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell wall modification | The series of events leading to chemical and structural alterations of an existing cell wall that can result in loosening, increased extensibility or disassembly. |
| pectin catabolic process | The chemical reactions and pathways resulting in the breakdown of pectin, a polymer containing a backbone of alpha-1,4-linked D-galacturonic acid residues. |
42 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3E8Z8 | PME28 | Putative pectinesterase/pectinesterase inhibitor 28 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GX86 | PME21 | Probable pectinesterase/pectinesterase inhibitor 21 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SRX4 | PME7 | Probable pectinesterase/pectinesterase inhibitor 7 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1JPL7 | PME18 | Pectinesterase/pectinesterase inhibitor 18 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84JX1 | PME19 | Probable pectinesterase/pectinesterase inhibitor 19 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49298 | PME6 | Probable pectinesterase/pectinesterase inhibitor 6 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q42534 | PME2 | Pectinesterase 2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q43867 | PME1 | Pectinesterase 1 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O48711 | PME12 | Probable pectinesterase/pectinesterase inhibitor 12 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q7Y201 | PME13 | Probable pectinesterase/pectinesterase inhibitor 13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SKX2 | PME16 | Probable pectinesterase/pectinesterase inhibitor 16 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22149 | PME17 | Probable pectinesterase/pectinesterase inhibitor 17 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O80722 | PME4 | Pectinesterase 4 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV8 | PME5 | Pectinesterase 5 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O22256 | PME20 | Probable pectinesterase/pectinesterase inhibitor 20 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M9W7 | PME22 | Putative pectinesterase/pectinesterase inhibitor 22 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8GXA1 | PME23 | Probable pectinesterase/pectinesterase inhibitor 23 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SG77 | PME24 | Putative pectinesterase/pectinesterase inhibitor 24 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O49006 | PME3 | Pectinesterase/pectinesterase inhibitor 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9STY3 | PME33 | Probable pectinesterase/pectinesterase inhibitor 33 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LYT5 | PME35 | Probable pectinesterase/pectinesterase inhibitor 35 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84R10 | PME36 | Probable pectinesterase/pectinesterase inhibitor 36 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q5MFV6 | VGDH2 | Probable pectinesterase/pectinesterase inhibitor VGDH2 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81320 | PME38 | Putative pectinesterase/pectinesterase inhibitor 38 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81415 | PME39 | Probable pectinesterase/pectinesterase inhibitor 39 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| O81301 | PME40 | Probable pectinesterase/pectinesterase inhibitor 40 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8RXK7 | PME41 | Probable pectinesterase/pectinesterase inhibitor 41 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q1PEC0 | PME42 | Probable pectinesterase/pectinesterase inhibitor 42 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY7 | PME44 | Probable pectinesterase/pectinesterase inhibitor 44 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9SMY6 | PME45 | Putative pectinesterase/pectinesterase inhibitor 45 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF78 | PME46 | Probable pectinesterase/pectinesterase inhibitor 46 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FF77 | PME47 | Probable pectinesterase/pectinesterase inhibitor 47 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXD9 | PME51 | Probable pectinesterase/pectinesterase inhibitor 51 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q3E989 | PME54 | Probable pectinesterase/pectinesterase inhibitor 54 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FJ21 | PME58 | Probable pectinesterase/pectinesterase inhibitor 58 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN5 | PME59 | Probable pectinesterase/pectinesterase inhibitor 59 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FHN4 | PME60 | Probable pectinesterase/pectinesterase inhibitor 60 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FK05 | PME61 | Probable pectinesterase/pectinesterase inhibitor 61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q8L7Q7 | PME64 | Probable pectinesterase/pectinesterase inhibitor 64 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q94CB1 | PME25 | Probable pectinesterase/pectinesterase inhibitor 25 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9LXK7 | PME32 | Probable pectinesterase/pectinesterase inhibitor 32 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M3B0 | PME34 | Probable pectinesterase/pectinesterase inhibitor 34 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MNKYVLLGVT | ALIMAMVICV | EANDGNSPSR | KMEEVHRESK | LMITKTTVSI | ICASTDYKQD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CTTSLATVRS | PDPRNLIRSA | FDLAIISIRS | GIDRGMIDLK | SRADADMHTR | EALNTCRELM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DDAIDDLRKT | RDKFRGFLFT | RLSDFVEDLC | VWLSGSITYQ | QTCIDGFEGI | DSEAAVMMER |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VMRKGQHLTS | NGLAIAANLD | KLLKAFRIPF | PFLRSRRGRL | GILGSGSSRD | ESVGSSQDSP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| PNEDSSDDSP | STVDSSENQP | VDSSSENQSS | DSSNNRPLDS | SKNQQMESSE | DTPKKSAFSG |
| 310 | 320 | 330 | 340 | 350 | 360 |
| NQPLDDSSDK | LPQKSTSSEN | QPLDSSENPP | QKSTSSENRP | LDPLRKLNPL | NKLDSLKDRH |
| 370 | 380 | 390 | 400 | 410 | 420 |
| LSEEGEFPPW | VTPHSRRLLA | RRPRNNGIKA | NVVVAKDGSG | KCKTIAQALA | MVPMKNTKKF |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VIHIKEGVYK | EKVEVTKKML | HVMFVGDGPT | KTVITGDIAF | LPDQVGTYRT | ASVAVNGDYF |
| 490 | 500 | 510 | 520 | 530 | 540 |
| MAKDIGFENT | AGAARHQAVA | LRVSADFAVF | FNCHMNGYQD | TLYVHTHRQF | YRNCRVSGTI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| DFVFGDAKAV | FQNCEFVIRR | PMEHQQCIVT | AQGRKDRRET | TGIVIHNSRI | TGDASYLPVK |
| 610 | 620 | 630 | 640 | 650 | 660 |
| AKNRAFLGRP | WKEFSRTIIM | NTEIDDVIDP | EGWLKWNETF | ALNTLFYTEY | RNRGRGSGQG |
| 670 | 680 | 690 | 700 | ||
| RRVRWRGIKR | ISDRAAREFA | PGNFLRGNTW | IPQTRIPYNA | N |