O15539
Gene name |
RGS5 |
Protein name |
Regulator of G-protein signaling 5 |
Names |
RGS5 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:8490 |
EC number |
|
Protein Class |
|
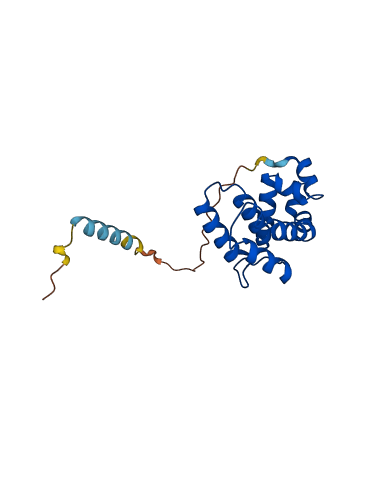
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

2 structures for O15539
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2CRP | NMR | - | A | 44-180 | PDB |
| AF-O15539-F1 | Predicted | AlphaFoldDB |
145 variants for O15539
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 4 | G>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs754269659 CA343466193 |
4 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1218846 rs754269659 |
4 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs867715021 CA31998105 |
6 | A>S | No |
ClinGen Ensembl |
|
|
CA343466182 rs1439478729 |
6 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs778098929 CA1218845 |
7 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA31998104 rs908862104 |
11 | S>* | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 13 | L>M | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 14 | E>D | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1161225071 CA343466134 |
14 | E>G | No |
ClinGen gnomAD |
|
|
CA1218840 rs762320638 |
14 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs753504362 CA1218817 |
16 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1276294311 CA343419838 |
16 | A>V | No |
ClinGen gnomAD |
|
|
rs760440634 CA1218815 |
18 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA343419800 rs1270633977 |
19 | I>T | No |
ClinGen gnomAD |
|
|
CA31847821 rs145871235 |
20 | K>N | No |
ClinGen ESP TOPMed |
|
|
rs775325062 CA1218814 |
21 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs1346060751 CA343419758 |
22 | K>N | No |
ClinGen gnomAD |
|
|
rs1571231174 CA343419698 |
27 | L>P | No |
ClinGen Ensembl |
|
| TCGA novel | 28 | Q>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA343419688 rs1282619058 |
28 | Q>R | No |
ClinGen gnomAD |
|
|
CA1218812 rs759505895 |
31 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA343419613 rs1300065405 |
34 | G>C | No |
ClinGen gnomAD |
|
|
CA343419564 rs1462700441 |
38 | I>F | No |
ClinGen gnomAD |
|
|
CA1218810 rs774429422 |
39 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA343419526 rs35968905 |
41 | N>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs35968905 CA1218808 |
41 | N>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1218807 rs374711151 |
41 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1474045884 CA343419512 |
42 | E>Q | No |
ClinGen gnomAD |
|
|
CA31847767 rs759477959 |
44 | P>A | No |
ClinGen gnomAD |
|
|
rs759477959 CA343419481 |
44 | P>S | No |
ClinGen gnomAD |
|
|
CA343419442 rs1253367758 |
47 | P>T | No |
ClinGen gnomAD |
|
|
rs1187670218 CA343419414 |
49 | K>T | No |
ClinGen TOPMed gnomAD |
|
|
rs747544052 CA1218785 |
53 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs1239832135 CA343419031 |
54 | S>L | No |
ClinGen gnomAD |
|
|
rs1183349685 CA343419035 |
54 | S>P | No |
ClinGen TOPMed |
|
|
rs777324995 CA343419020 |
56 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1218782 rs748874557 |
56 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755737266 CA1218780 |
57 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 57 | E>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1218779 rs752330335 |
58 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767246365 CA1218778 |
62 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA1218777 rs754717880 |
62 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1380159437 CA343418979 |
63 | D>H | No |
ClinGen TOPMed |
|
|
CA343418967 rs1324830436 |
64 | S>Y | No |
ClinGen gnomAD |
|
|
CA31843105 rs369125258 |
65 | L>P | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1452385815 CA343418955 |
66 | D>E | No |
ClinGen gnomAD |
|
|
rs1251394136 CA343418962 |
66 | D>N | No |
ClinGen TOPMed |
|
|
rs1361587373 CA343418944 |
68 | L>F | No |
ClinGen gnomAD |
|
|
rs751332579 CA1218776 |
70 | Q>R | No |
ClinGen ExAC |
|
| TCGA novel | 72 | N>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1044247757 CA31843097 |
72 | N>S | No |
ClinGen TOPMed |
|
|
rs750306051 CA1218754 |
73 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
COSM400882 CA343418117 rs866971353 |
74 | G>* | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
COSM1663917 rs866971353 CA31837898 |
74 | G>R | kidney [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1018911732 CA31837892 |
77 | S>N | No |
ClinGen Ensembl |
|
|
rs1404998460 CA343418033 |
79 | K>E | No |
ClinGen gnomAD |
|
|
rs375964318 CA1218752 |
79 | K>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA343417995 rs1557880075 |
80 | S>N | No |
ClinGen Ensembl |
|
|
CA343417983 rs753832247 |
80 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA343417991 rs1557880075 |
80 | S>T | No |
ClinGen Ensembl |
|
|
CA1218749 rs201830771 |
87 | S>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA31837872 rs1027034349 |
88 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1266978669 CA343417765 |
90 | N>D | No |
ClinGen TOPMed |
|
|
CA1218747 rs143682816 |
90 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 90 | N>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs200762280 CA1218745 |
92 | E>D | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA1218746 rs772501697 |
92 | E>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 93 | F>L | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1036136629 CA31837856 |
94 | W>R | No |
ClinGen TOPMed gnomAD |
|
|
CA1218744 rs148930012 |
95 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA343417648 rs1251592440 |
97 | C>F | No |
ClinGen gnomAD |
|
|
rs1251592440 CA343417650 |
97 | C>Y | No |
ClinGen gnomAD |
|
|
CA1218743 rs771316827 |
99 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs1316489572 CA343417600 |
100 | Y>D | No |
ClinGen gnomAD |
|
|
CA1218742 rs747730118 |
102 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA343417547 rs1386893999 |
103 | I>N | No |
ClinGen TOPMed |
|
|
CA31837840 rs906463602 |
104 | K>R | No |
ClinGen Ensembl |
|
|
rs141283960 CA1218741 |
105 | S>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1392771881 CA343417476 |
108 | K>E | No |
ClinGen TOPMed |
|
|
rs1280808730 CA343417452 |
109 | M>I | No |
ClinGen gnomAD |
|
| TCGA novel | 110 | A>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs746606578 CA1218738 |
110 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1218737 rs779826661 |
110 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1334596366 CA343417438 |
112 | K>E | No |
ClinGen TOPMed |
|
|
rs190048374 CA1218736 |
112 | K>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA343417409 rs1389428885 |
116 | I>V | No |
ClinGen gnomAD |
|
|
rs757138090 CA343417397 |
117 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs144088608 CA1218734 |
117 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs144088608 CA343417399 |
117 | Y>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1218732 rs754008466 |
122 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764215537 CA343417355 |
123 | T>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764215537 CA1218731 |
123 | T>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs768014429 CA1218728 |
125 | A>P | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 125 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA1218727 rs759991833 |
127 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755146862 CA1218707 |
129 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA1218706 rs752015607 |
131 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs1557877821 CA343415973 |
132 | D>G | No |
ClinGen Ensembl |
|
|
rs1291024162 CA343415906 |
135 | T>A | No |
ClinGen TOPMed |
|
|
rs1315372347 CA343415852 |
137 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA343415840 rs1434786120 |
137 | D>V | No |
ClinGen TOPMed |
|
|
rs766595377 CA1218705 |
138 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA31835278 rs1052965505 |
140 | M>R | No |
ClinGen Ensembl |
|
|
CA343415774 rs1295762998 |
140 | M>V | No |
ClinGen TOPMed |
|
|
CA1218704 rs148613695 |
141 | K>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA1218703 rs773667539 |
142 | N>S | No |
ClinGen ExAC gnomAD |
|
|
rs763615294 CA1218702 |
144 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs1367658247 CA343415597 |
146 | P>H | No |
ClinGen gnomAD |
|
|
rs760220434 CA1218701 |
146 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA1218700 rs774978465 |
147 | S>F | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1412104524 CA343415543 |
148 | L>R | No |
ClinGen gnomAD |
|
| TCGA novel | 149 | S>G | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA343415526 rs1476025671 |
149 | S>N | No |
ClinGen gnomAD |
|
|
rs1311567901 CA343415541 |
149 | S>R | No |
ClinGen TOPMed |
|
|
CA31835258 rs1007594231 |
149 | S>R | No |
ClinGen TOPMed |
|
|
rs1262386674 CA343415484 |
151 | F>S | No |
ClinGen TOPMed |
|
|
CA1218697 rs774205323 |
153 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs745454274 CA1218698 |
153 | M>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA343415391 rs1242366596 |
155 | Q>E | No |
ClinGen Ensembl |
|
|
rs1490710140 CA343415386 |
155 | Q>R | No |
ClinGen gnomAD |
|
|
CA343415366 rs1477897678 |
156 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
CA31835244 rs770595330 |
157 | R>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA1218696 rs770595330 |
157 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA343415315 rs1247708336 |
158 | I>S | No |
ClinGen gnomAD |
|
|
rs1221312842 CA343415305 |
159 | H>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs777909194 CA1218694 |
160 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA1218693 rs756084760 |
162 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA1218692 rs748302072 |
164 | K>E | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 164 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 165 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA343415139 rs1398193231 |
166 | S>P | No |
ClinGen gnomAD |
|
|
CA1218690 rs755241558 |
167 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA1218688 rs751665168 |
168 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA1218687 rs192189511 |
169 | R>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs758639664 CA1218686 COSM1335818 |
169 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 170 | F>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1205978249 CA343415000 |
171 | V>G | No |
ClinGen gnomAD |
|
|
CA1218684 rs1457102023 |
171 | V>L | No |
ClinGen TOPMed |
|
|
COSM369226 CA1218683 rs112323155 |
172 | R>C | lung kidney [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
COSM530240 rs147130564 CA1218682 |
172 | R>H | lung [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA31835186 rs147130564 |
172 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA343414922 rs1571205111 |
175 | F>V | No |
ClinGen Ensembl |
|
|
CA1218681 rs762295521 |
182 | K>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA1218680 rs774994927 |
182 | K>W | No |
ClinGen ExAC gnomAD |
No associated diseases with O15539
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTPase activator activity | Binds to and increases the activity of a GTPase, an enzyme that catalyzes the hydrolysis of GTP. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| G protein-coupled receptor signaling pathway | The series of molecular signals initiated by a ligand binding to its receptor, in which the activated receptor promotes the exchange of GDP for GTP on the alpha-subunit of an associated heterotrimeric G-protein complex. The GTP-bound activated alpha-G-protein then dissociates from the beta- and gamma-subunits to further transmit the signal within the cell. The pathway begins with receptor-ligand interaction, and ends with regulation of a downstream cellular process. The pathway can start from the plasma membrane, Golgi or nuclear membrane. |
| negative regulation of signal transduction | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction. |
| regulation of G protein-coupled receptor signaling pathway | Any process that modulates the frequency, rate or extent of G protein-coupled receptor signaling pathway. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| O46471 | RGS16 | Regulator of G-protein signaling 16 | Bos taurus (Bovine) | PR |
| Q3T0T8 | RGS5 | Regulator of G-protein signaling 5 | Bos taurus (Bovine) | PR |
| P49802 | RGS7 | Regulator of G-protein signaling 7 | Homo sapiens (Human) | PR |
| Q9Z2H1 | Rgs11 | Regulator of G-protein signaling 11 | Mus musculus (Mouse) | PR |
| O54829 | Rgs7 | Regulator of G-protein signaling 7 | Mus musculus (Mouse) | PR |
| Q0QWG9 | Grid2ip | Delphilin | Mus musculus (Mouse) | PR |
| Q18312 | rgs-3 | Regulator of G-protein signaling rgs-3 | Caenorhabditis elegans | PR |
| Q8H1F2 | RGS1 | Regulator of G-protein signaling 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MCKGLAALPH | SCLERAKEIK | IKLGILLQKP | DSVGDLVIPY | NEKPEKPAKT | QKTSLDEALQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| WRDSLDKLLQ | NNYGLASFKS | FLKSEFSEEN | LEFWIACEDY | KKIKSPAKMA | EKAKQIYEEF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IQTEAPKEVN | IDHFTKDITM | KNLVEPSLSS | FDMAQKRIHA | LMEKDSLPRF | VRSEFYQELI |
| K |