O15498
Gene name |
YKT6 |
Protein name |
Synaptobrevin homolog YKT6 |
Names |
EC 2.3.1.- |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:10652 |
EC number |
2.3.1.-: Transferring groups other than amino-acyl groups |
Protein Class |
|
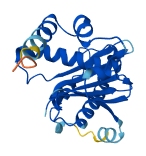
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
138-198 (v-SNARE coiled-coil homology) |
Relief mechanism |
PTM |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

4 structures for O15498
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 6J74 | X-ray | 321 A | C | 1-198 | PDB |
| 6J7F | X-ray | 288 A | C | 1-195 | PDB |
| 6J7X | X-ray | 275 A | C | 1-198 | PDB |
| AF-O15498-F1 | Predicted | AlphaFoldDB |
173 variants for O15498
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| TCGA novel | 2 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1562740487 | 2 | K>Q | No | Ensembl | |
| rs2096335005 | 2 | K>R | No | TOPMed | |
| rs957904298 | 3 | L>P | No |
TOPMed gnomAD |
|
| rs557185462 | 3 | L>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2096335019 | 4 | Y>H | No | gnomAD | |
| rs757356224 | 5 | S>R | No |
ExAC gnomAD |
|
| rs1278676175 | 5 | S>T | No | gnomAD | |
| rs2096335034 | 6 | L>F | No | Ensembl | |
| rs778906573 | 7 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1460636013 | 8 | V>G | No | gnomAD | |
| rs141044177 | 9 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs141044177 | 9 | L>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1444571069 | 10 | Y>* | No | gnomAD | |
| rs150272603 | 10 | Y>C | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1188872212 | 11 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2096335075 | 12 | G>A | No | TOPMed | |
| rs2096335087 | 13 | E>K | No | Ensembl | |
| rs1260480876 | 14 | A>S | No |
TOPMed gnomAD |
|
| rs2096335094 | 15 | K>E | No | Ensembl | |
| rs755355202 | 16 | V>M | No |
ExAC gnomAD |
|
| rs748870 | 17 | V>G | No | Ensembl | |
| rs1486529076 | 18 | L>Q | No |
TOPMed gnomAD |
|
| rs2096335125 | 19 | L>F | No | TOPMed | |
| rs1562740562 | 20 | K>R | No | Ensembl | |
| rs866184383 | 21 | A>S | No | Ensembl | |
| rs774022853 | 22 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs774022853 | 22 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs2096335153 | 22 | A>V | No | Ensembl | |
|
COSM3431616 rs374169220 |
23 | Y>* | Variant assessed as Somatic; HIGH impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ESP ExAC TOPMed gnomAD |
| rs745479059 | 23 | Y>C | No |
ExAC TOPMed gnomAD |
|
| rs775371239 | 24 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs147946572 | 25 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs763866140 | 26 | S>F | No |
ExAC gnomAD |
|
| rs776602646 | 27 | S>A | No |
ExAC TOPMed gnomAD |
|
| rs2096335183 | 27 | S>F | No | TOPMed | |
| rs776602646 | 27 | S>P | No |
ExAC TOPMed gnomAD |
|
| rs370881857 | 28 | F>L | No |
ESP ExAC gnomAD |
|
| rs750502766 | 29 | S>N | No |
ExAC gnomAD |
|
| rs750502766 | 29 | S>T | No |
ExAC gnomAD |
|
| rs2128837798 | 32 | Q>E | No | Ensembl | |
| rs2096335201 | 33 | R>K | No | Ensembl | |
| rs2096335205 | 34 | S>F | No | gnomAD | |
| rs200537015 | 36 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs374571498 | 36 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
|
rs374571498 COSM3785698 |
36 | V>I | pancreas [Cosmic] | No |
cosmic curated ESP ExAC TOPMed gnomAD |
| rs2096338959 | 39 | F>V | No | Ensembl | |
| rs185914544 | 40 | M>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1367857316 | 41 | T>I | No | gnomAD | |
| rs753008649 | 43 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs753008649 | 43 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1265224121 | 44 | S>G | No | TOPMed | |
| rs1366763391 | 44 | S>N | No | gnomAD | |
| rs778414993 | 47 | I>L | No |
ExAC gnomAD |
|
| rs754467340 | 47 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1359419247 | 47 | I>T | No | gnomAD | |
| rs1267675271 | 49 | E>Q | No | gnomAD | |
| rs2096339012 | 49 | E>V | No | TOPMed | |
| rs199843184 | 50 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs199843184 | 50 | R>G | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs368134203 | 50 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs141563755 | 52 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs141563755 | 52 | S>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs967331681 | 55 | T>A | No |
TOPMed gnomAD |
|
| rs1384514874 | 55 | T>I | No | gnomAD | |
| rs780632333 | 57 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1286657456 | 60 | K>E | No | gnomAD | |
| rs769642918 | 60 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs2096339073 | 62 | Q>H | No | gnomAD | |
| rs138273846 | 64 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1446402208 | 64 | Y>H | No | gnomAD | |
| rs138273846 | 64 | Y>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs143932038 | 66 | C>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs1160617543 | 67 | H>Y | No |
TOPMed gnomAD |
|
|
COSM3768392 rs376107091 |
68 | V>I | liver [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs372618652 | 69 | Y>C | No |
ESP ExAC gnomAD |
|
| rs1277939353 | 70 | V>I | No |
TOPMed gnomAD |
|
| rs761029409 | 71 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
rs775548288 COSM1233022 |
71 | R>W | large_intestine [Cosmic] | No |
cosmic curated ExAC TOPMed gnomAD |
| rs202098672 | 72 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs1480300991 COSM223155 |
74 | S>N | skin [Cosmic] | No |
cosmic curated TOPMed gnomAD |
| rs754256887 | 82 | D>V | No |
ExAC gnomAD |
|
| rs2096340996 | 86 | P>R | No | TOPMed | |
| rs2096340987 | 86 | P>T | No | Ensembl | |
| rs765644782 | 88 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs765644782 | 88 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs550011837 | 88 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs754414111 | 89 | V>L | No |
ExAC gnomAD |
|
| rs2096341025 | 90 | A>T | No | TOPMed | |
| rs767101133 | 90 | A>V | No |
ExAC gnomAD |
|
| rs752296251 | 94 | L>Q | No |
ExAC gnomAD |
|
| rs2096341055 | 95 | E>A | No | gnomAD | |
| rs571774649 | 95 | E>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2128839394 | 96 | K>* | No | Ensembl | |
| rs1354516091 | 99 | D>E | No |
TOPMed gnomAD |
|
| rs1429614754 | 101 | F>C | No | gnomAD | |
| TCGA novel | 101 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs143284758 | 102 | S>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs773570225 | 102 | S>P | No |
ExAC gnomAD |
|
| rs773570225 | 102 | S>T | No |
ExAC gnomAD |
|
| rs1323377543 | 105 | V>A | No | gnomAD | |
|
rs1438883980 COSM3431617 |
106 | D>N | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated NCI-TCGA gnomAD |
| rs1473695508 | 107 | R>S | No |
TOPMed gnomAD |
|
| rs1014962983 | 110 | W>* | No |
TOPMed gnomAD |
|
| rs755707761 | 110 | W>R | No |
ExAC TOPMed gnomAD |
|
| rs1014962983 | 110 | W>S | No |
TOPMed gnomAD |
|
|
rs1583647564 COSM1089929 |
111 | P>S | Variant assessed as Somatic; MODERATE impact. endometrium [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated Ensembl |
| rs1583647572 | 112 | V>G | No | Ensembl | |
| rs1226032982 | 113 | G>A | No | gnomAD | |
| rs2096341939 | 113 | G>R | No | TOPMed | |
| rs1299696747 | 114 | S>A | No | gnomAD | |
| rs112426773 | 114 | S>C | No | Ensembl | |
| rs1583647600 | 115 | P>S | No | Ensembl | |
| TCGA novel | 118 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs963366243 | 118 | I>V | No | TOPMed | |
| rs1583647615 | 119 | H>P | No | Ensembl | |
| rs1209010661 | 120 | Y>F | No | gnomAD | |
| rs2096341973 | 121 | P>L | No | gnomAD | |
| rs2096341975 | 122 | A>V | No | Ensembl | |
| rs144641826 | 123 | L>V | No |
1000Genomes ExAC gnomAD |
|
| rs570507763 | 124 | D>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2096341981 | 124 | D>N | No | Ensembl | |
| rs201188604 | 126 | H>L | No | 1000Genomes | |
| rs1321161946 | 126 | H>Q | No |
TOPMed gnomAD |
|
| TCGA novel | 129 | R>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2096342001 | 129 | R>S | No | TOPMed | |
| rs1246829336 | 130 | Y>H | No | gnomAD | |
| rs959201807 | 131 | Q>* | No | Ensembl | |
|
COSM4741858 rs753469894 |
134 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs2096342920 | 134 | R>Q | No | TOPMed | |
| TCGA novel | 135 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs761553542 | 136 | A>T | No |
ExAC gnomAD |
|
| rs764904707 | 136 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1266027342 | 137 | D>H | No | TOPMed | |
| rs1019274884 | 139 | M>T | No | TOPMed | |
| rs1206624926 | 139 | M>V | No | TOPMed | |
| rs750242578 | 140 | T>I | No |
ExAC TOPMed gnomAD |
|
|
rs780133803 COSM172290 |
145 | E>K | Variant assessed as Somatic; MODERATE impact. large_intestine [NCI-TCGA, Cosmic] | No |
NCI-TCGA Cosmic cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| rs2096342976 | 148 | E>G | No | Ensembl | |
| rs772944817 | 150 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs772944817 | 150 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs2096342982 | 151 | I>V | No | TOPMed | |
| rs762715210 | 155 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs2096346244 | 156 | T>S | No | Ensembl | |
| rs774293163 | 158 | E>G | No |
ExAC gnomAD |
|
| rs1488272305 | 158 | E>K | No | gnomAD | |
| rs767550148 | 159 | S>A | No |
ExAC gnomAD |
|
| rs752644714 | 160 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs756189723 | 162 | E>G | No |
ExAC gnomAD |
|
| COSM1089930 | 163 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2096346263 | 163 | R>G | No | Ensembl | |
| rs764130626 | 163 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1170871324 | 164 | G>A | No | gnomAD | |
| rs754045609 | 165 | E>G | No |
ExAC gnomAD |
|
| rs2096346284 | 171 | V>M | No | Ensembl | |
| rs866811977 | 172 | S>F | No | Ensembl | |
| rs2096346303 | 175 | E>K | No | Ensembl | |
| rs916832961 | 176 | V>L | No |
TOPMed gnomAD |
|
| rs2128840745 | 180 | Q>* | No | Ensembl | |
| COSM1313106 | 181 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1363641420 | 182 | K>* | No | gnomAD | |
| rs2096346323 | 183 | A>V | No | TOPMed | |
| COSM1089931 | 186 | K>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1583650537 | 187 | T>P | No | Ensembl | |
| rs747509605 | 189 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs141713796 | 189 | R>Q | No |
ESP TOPMed gnomAD |
|
| rs747509605 | 189 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1183379380 | 190 | K>E | No | gnomAD | |
| rs2096347668 | 193 | S>A | No | Ensembl | |
| rs2096347669 | 194 | C>R | No | Ensembl | |
| rs774116833 | 194 | C>W | No |
ExAC TOPMed gnomAD |
|
| rs1174763015 | 196 | A>V | No | gnomAD | |
| rs1359023676 | 197 | I>V | No | gnomAD |
No associated diseases with O15498
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.1.- | Transferring groups other than amino-acyl groups |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
15 GO annotations of cellular component
| Name | Definition |
|---|---|
| apical dendrite | A dendrite that emerges near the apical pole of a neuron. In bipolar neurons, apical dendrites are located on the opposite side of the soma from the axon. |
| basal dendrite | A dendrite that emerges near the basal pole of a neuron. In bipolar neurons, basal dendrites are either on the same side of the soma as the axon, or project toward the axon. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic vesicle membrane | The lipid bilayer surrounding a cytoplasmic vesicle. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| endoplasmic reticulum | The irregular network of unit membranes, visible only by electron microscopy, that occurs in the cytoplasm of many eukaryotic cells. The membranes form a complex meshwork of tubular channels, which are often expanded into slitlike cavities called cisternae. The ER takes two forms, rough (or granular), with ribosomes adhering to the outer surface, and smooth (with no ribosomes attached). |
| endoplasmic reticulum-Golgi intermediate compartment membrane | The lipid bilayer surrounding any of the compartments of the endoplasmic reticulum (ER)-Golgi intermediate compartment system. |
| endosome | A vacuole to which materials ingested by endocytosis are delivered. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| Golgi membrane | The lipid bilayer surrounding any of the compartments of the Golgi apparatus. |
| mitochondrion | A semiautonomous, self replicating organelle that occurs in varying numbers, shapes, and sizes in the cytoplasm of virtually all eukaryotic cells. It is notably the site of tissue respiration. |
| neuronal cell body | The portion of a neuron that includes the nucleus, but excludes cell projections such as axons and dendrites. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| SNARE complex | A protein complex involved in membrane fusion; a stable ternary complex consisting of a four-helix bundle, usually formed from one R-SNARE and three Q-SNAREs with an ionic layer sandwiched between hydrophobic layers. One well-characterized example is the neuronal SNARE complex formed of synaptobrevin 2, syntaxin 1a, and SNAP-25. |
| transport vesicle | Any of the vesicles of the constitutive secretory pathway, which carry cargo from the endoplasmic reticulum to the Golgi, between Golgi cisternae, from the Golgi to the ER (retrograde transport) or to destinations within or outside the cell. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| protein-cysteine S-palmitoyltransferase activity | Catalysis of the transfer of a palmitoyl (systematic name, hexadecanoyl) group to a sulfur atom on the cysteine of a protein molecule, in the reaction hexadecanoyl-CoA + L-cysteinyl- = CoA + S-hexadecanoyl-L-cysteinyl-. |
| SNAP receptor activity | Acting as a marker to identify a membrane and interacting selectively with one or more SNAREs on another membrane to mediate membrane fusion. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| endoplasmic reticulum to Golgi vesicle-mediated transport | The directed movement of substances from the endoplasmic reticulum (ER) to the Golgi, mediated by COP II vesicles. Small COP II coated vesicles form from the ER and then fuse directly with the cis-Golgi. Larger structures are transported along microtubules to the cis-Golgi. |
| protein transport | The directed movement of proteins into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| retrograde transport, endosome to Golgi | The directed movement of membrane-bounded vesicles from endosomes back to the trans-Golgi network where they are recycled for further rounds of transport. |
| vesicle docking involved in exocytosis | The initial attachment of a vesicle membrane to a target membrane, mediated by proteins protruding from the membrane of the vesicle and the target membrane, that contributes to exocytosis. |
| vesicle targeting | The process in which vesicles are directed to specific destination membranes. Targeting involves coordinated interactions among cytoskeletal elements (microtubules or actin filaments), motor proteins, molecules at the vesicle membrane and target membrane surfaces, and vesicle cargo. |
8 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P36015 | YKT6 | Synaptobrevin homolog YKT6 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | EV |
| Q3T000 | YKT6 | Synaptobrevin homolog YKT6 | Bos taurus (Bovine) | SS |
| Q9CQW1 | Ykt6 | Synaptobrevin homolog YKT6 | Mus musculus (Mouse) | SS |
| Q5EGY4 | Ykt6 | Synaptobrevin homolog YKT6 | Rattus norvegicus (Rat) | EV |
| Q9LVM9 | YKT62 | VAMP-like protein YKT62 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZRD6 | YKT61 | VAMP-like protein YKT61 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q6P816 | ykt6 | Synaptobrevin homolog YKT6 | Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) | SS |
| Q7ZUN8 | ykt6 | Synaptobrevin homolog YKT6 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKLYSLSVLY | KGEAKVVLLK | AAYDVSSFSF | FQRSSVQEFM | TFTSQLIVER | SSKGTRASVK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EQDYLCHVYV | RNDSLAGVVI | ADNEYPSRVA | FTLLEKVLDE | FSKQVDRIDW | PVGSPATIHY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PALDGHLSRY | QNPREADPMT | KVQAELDETK | IILHNTMESL | LERGEKLDDL | VSKSEVLGTQ |
| 190 | |||||
| SKAFYKTARK | QNSCCAIM |