O15031
Gene name |
PLXNB2 (KIAA0315) |
Protein name |
Plexin-B2 |
Names |
MM1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:23654 |
EC number |
|
Protein Class |
|
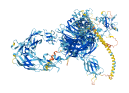
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
1271-1805 (Plexin, cytoplasmic RasGAP domain) |
Relief mechanism |
Partner binding |
Assay |
|
Target domain |
1271-1805 (Plexin, cytoplasmic RasGAP domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Wang Y et al. (2012) "Plexins are GTPase-activating proteins for Rap and are activated by induced dimerization", Science signaling, 5, ra6
- He H et al. (2009) "Crystal structure of the plexin A3 intracellular region reveals an autoinhibited conformation through active site sequestration", Proceedings of the National Academy of Sciences of the United States of America, 106, 15610-5
Autoinhibited structure

Activated structure

2 structures for O15031
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4E71 | X-ray | 226 A | A | 1452-1562 | PDB |
| AF-O15031-F1 | Predicted | AlphaFoldDB |
1857 variants for O15031
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs1471417012 | 2 | A>T | No |
TOPMed gnomAD |
|
| rs542502824 | 4 | Q>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs755797039 | 5 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs755797039 | 5 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs755797039 | 5 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs2066794331 | 7 | A>T | No | TOPMed | |
| rs752601544 | 7 | A>V | No |
ExAC gnomAD |
|
| rs1459764825 | 9 | T>I | No |
TOPMed gnomAD |
|
| rs1328015651 | 9 | T>P | No | gnomAD | |
| rs1328015651 | 9 | T>S | No | gnomAD | |
| rs907874054 | 11 | L>V | No | TOPMed | |
| rs1370982481 | 13 | L>P | No | gnomAD | |
| rs1476297870 | 13 | L>V | No | gnomAD | |
|
COSM4105109 rs372428698 |
16 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs761933126 | 16 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs764370385 | 17 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs764370385 | 17 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs954590766 | 18 | A>D | No | gnomAD | |
| rs954590766 | 18 | A>V | No | gnomAD | |
| rs2147537152 | 19 | S>I | No | Ensembl | |
| rs2066790896 | 20 | L>P | No | Ensembl | |
| rs2066790742 | 21 | R>K | No | Ensembl | |
| rs775938208 | 21 | R>S | No |
ExAC TOPMed gnomAD |
|
|
COSM3694168 rs546928788 |
23 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs370518066 | 23 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs774670910 | 24 | K>E | No |
ExAC gnomAD |
|
| TCGA novel | 25 | L>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066788765 | 26 | D>A | No | TOPMed | |
| TCGA novel | 26 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748847916 | 28 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs777413113 | 29 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs769625156 | 29 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs769625156 | 29 | R>L | No |
ExAC TOPMed gnomAD |
|
| COSM479118 | 31 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs747741923 COSM1417085 |
31 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs199944521 | 32 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs2066787228 | 33 | E>V | No |
TOPMed gnomAD |
|
| rs2066786554 | 34 | L>P | No | Ensembl | |
| rs1171178234 | 36 | H>L | No | gnomAD | |
| rs1171178234 | 36 | H>P | No | gnomAD | |
| rs1252912952 | 37 | L>P | No | gnomAD | |
| rs758332139 | 39 | V>G | No |
ExAC gnomAD |
|
| rs1248795262 | 41 | E>D | No | gnomAD | |
| rs372801431 | 42 | A>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1569175477 | 43 | S>L | No | Ensembl | |
| rs756335548 | 44 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1347160019 | 45 | V>A | No | gnomAD | |
| rs201675007 | 45 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1280225940 | 46 | V>L | No |
TOPMed gnomAD |
|
| rs1376885639 | 50 | A>S | No |
TOPMed gnomAD |
|
| rs774745979 | 50 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs923222375 | 51 | V>L | No | gnomAD | |
| rs923222375 | 51 | V>M | No | gnomAD | |
| rs2066780796 | 53 | A>T | No | Ensembl | |
| rs772859230 | 54 | L>V | No |
ExAC gnomAD |
|
| rs1471417535 | 55 | Y>* | No | gnomAD | |
| rs1169738620 | 56 | Q>H | No | Ensembl | |
| rs747863495 | 57 | L>M | No |
ExAC gnomAD |
|
| rs1181507364 | 59 | A>T | No | gnomAD | |
| rs776198723 | 59 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs375360559 | 62 | Q>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2147535142 | 62 | Q>H | No | Ensembl | |
| rs2147535179 | 62 | Q>L | No | Ensembl | |
| TCGA novel | 63 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs536602240 | 64 | E>K | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1601729637 | 67 | V>G | No | Ensembl | |
| rs758346334 | 67 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1225579986 | 68 | A>T | No | gnomAD | |
| rs2066777898 | 68 | A>V | No |
TOPMed gnomAD |
|
| rs745740476 | 69 | T>K | No |
ExAC TOPMed gnomAD |
|
| rs745740476 | 69 | T>M | No |
ExAC TOPMed gnomAD |
|
|
COSM3065409 rs1233673658 |
71 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1212847385 | 72 | A>G | No | TOPMed | |
| rs1212847385 | 72 | A>V | No | TOPMed | |
| rs1310856490 | 73 | L>V | No |
TOPMed gnomAD |
|
| rs1413731587 | 74 | D>E | No |
TOPMed gnomAD |
|
| rs767671854 | 76 | K>N | No |
ExAC gnomAD |
|
| rs2147534649 | 76 | K>Q | No | Ensembl | |
| rs1349167729 | 76 | K>R | No | Ensembl | |
| TCGA novel | 77 | K>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2147534562 | 77 | K>* | No | Ensembl | |
| TCGA novel | 77 | K>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs569631269 | 79 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM726727 | 80 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1475828872 | 81 | P>R | No | gnomAD | |
| rs773812866 | 82 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1420414767 | 82 | I>V | No | gnomAD | |
| rs1474985932 | 84 | A>P | No | gnomAD | |
| rs2066774409 | 85 | S>G | No |
TOPMed gnomAD |
|
| TCGA novel | 85 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs765819450 | 86 | Q>E | No |
ExAC gnomAD |
|
| rs1218028526 | 88 | H>Y | No | gnomAD | |
| rs1283660890 | 90 | A>V | No | gnomAD | |
| rs1555925101 | 91 | E>Q | No | Ensembl | |
| rs1243486713 | 92 | M>I | No | TOPMed | |
| rs376633792 | 94 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs768254955 | 95 | N>S | No |
ExAC gnomAD |
|
| rs1569175137 | 99 | L>Q | No | Ensembl | |
| rs367645843 | 102 | L>F | No |
ESP ExAC |
|
| COSM4164977 | 103 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2147533807 | 103 | D>Y | No | Ensembl | |
| rs778869284 | 105 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs1322525284 | 107 | K>N | No |
TOPMed gnomAD |
|
| rs1397375327 | 107 | K>R | No | gnomAD | |
| rs2066770846 | 108 | R>C | No | TOPMed | |
| rs770869553 | 108 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs770869553 | 108 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs2066770846 | 108 | R>S | No | TOPMed | |
| rs1405202944 | 110 | V>M | No | gnomAD | |
| TCGA novel | 111 | E>Q | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1601729395 | 112 | C>G | No | Ensembl | |
| rs200827623 | 113 | G>A | No |
ExAC gnomAD |
|
| rs200827623 | 113 | G>D | No |
ExAC gnomAD |
|
| rs1474555294 | 113 | G>S | No | gnomAD | |
| rs1006977673 | 114 | S>R | No | Ensembl | |
| rs1194371646 | 114 | S>T | No | gnomAD | |
| rs1601729342 | 117 | K>Q | No | Ensembl | |
| rs2147533272 | 119 | I>T | No | Ensembl | |
| rs755165035 | 121 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs755165035 | 121 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1176764896 | 121 | A>V | No | Ensembl | |
| rs751734137 | 122 | L>Q | No |
ExAC gnomAD |
|
| TCGA novel | 122 | L>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1307927339 | 123 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1344834441 | 123 | R>H | No |
TOPMed gnomAD |
|
| rs766516184 | 124 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs766516184 | 124 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs1360874782 | 125 | L>Q | No | gnomAD | |
| rs1303463814 | 126 | S>N | No |
TOPMed gnomAD |
|
| rs750788046 | 126 | S>R | No |
ExAC gnomAD |
|
| rs2066766735 | 127 | N>H | No |
TOPMed gnomAD |
|
| rs765914980 | 128 | I>V | No |
ExAC gnomAD |
|
| rs762372626 | 129 | S>C | No |
ExAC gnomAD |
|
| COSM3555709 | 129 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1436402793 | 130 | L>F | No |
TOPMed gnomAD |
|
| rs753306834 | 130 | L>H | No |
ExAC TOPMed gnomAD |
|
| rs753306834 | 130 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs756123018 | 131 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs756123018 | 131 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs368491965 | 131 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs368491965 | 131 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| COSM4105107 | 134 | Y>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs774126870 COSM1417084 |
135 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs200224862 | 136 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs749186574 | 137 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs781132198 | 138 | S>N | No |
ExAC TOPMed gnomAD |
|
| COSM4105106 | 139 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1035420 | 140 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM1035421 | 140 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4414567 | 141 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM4414571 | 141 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
rs2066764509 COSM1035419 |
144 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic Ensembl |
| rs780465304 | 145 | A>T | No |
ExAC gnomAD |
|
| rs2066763762 | 148 | D>N | No | Ensembl | |
| COSM461055 | 149 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs368559599 | 150 | G>D | No |
ESP ExAC TOPMed gnomAD |
|
| rs374494862 | 151 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs374494862 | 151 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1281599374 | 154 | V>A | No |
TOPMed gnomAD |
|
| rs2066762960 | 155 | G>E | No | gnomAD | |
| rs1386653254 | 156 | L>R | No | gnomAD | |
| rs1601729143 | 157 | V>G | No | Ensembl | |
| rs2066762124 | 158 | S>N | No | TOPMed | |
| rs1440523203 | 159 | S>F | No |
TOPMed gnomAD |
|
| rs763537846 | 160 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs767220661 | 163 | G>D | No |
ExAC gnomAD |
|
| rs767220661 | 163 | G>V | No |
ExAC gnomAD |
|
| rs1410136351 | 164 | G>D | No |
TOPMed gnomAD |
|
| rs774141104 | 164 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs201610707 | 166 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs201610707 | 166 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs199977078 | 166 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs201610707 | 166 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs775699976 | 167 | V>L | No |
ExAC TOPMed gnomAD |
|
|
rs775699976 COSM4105105 |
167 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs1484500987 | 170 | V>M | No | gnomAD | |
| rs772437014 | 174 | N>D | No |
ExAC gnomAD |
|
| rs1318021803 | 174 | N>S | No |
TOPMed gnomAD |
|
| TCGA novel | 175 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1222418641 | 176 | P>L | No | gnomAD | |
| rs779180914 | 177 | H>D | No |
ExAC TOPMed gnomAD |
|
| rs370376989 | 177 | H>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs779180914 | 177 | H>Y | No |
ExAC TOPMed gnomAD |
|
| rs1033777280 | 179 | N>D | No | gnomAD | |
| rs201664331 | 179 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs373060838 | 180 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs751172888 | 183 | V>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 184 | S>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1397038623 | 185 | T>S | No | Ensembl | |
|
rs531070913 COSM5393895 |
186 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 189 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs377754571 | 190 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs765277978 | 190 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs765277978 | 190 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs377754571 | 190 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| rs761785573 | 191 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs775716596 | 192 | D>N | No |
ExAC gnomAD |
|
| rs1248287051 | 193 | S>G | No |
TOPMed gnomAD |
|
| rs1017134736 | 193 | S>N | No | TOPMed | |
| rs2066755387 | 194 | R>S | No |
TOPMed gnomAD |
|
| TCGA novel | 194 | R>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374426305 | 195 | E>D | No |
ESP ExAC gnomAD |
|
| rs2147530555 | 196 | A>V | No | Ensembl | |
| rs1294258423 | 200 | Y>C | No |
TOPMed gnomAD |
|
| rs1294258423 | 200 | Y>F | No |
TOPMed gnomAD |
|
| rs368995795 | 200 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs1007052945 | 201 | T>K | No |
TOPMed gnomAD |
|
| rs1007052945 | 201 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1601728895 | 202 | D>A | No | Ensembl | |
| rs563687885 | 203 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777982464 | 204 | A>T | No |
ExAC gnomAD |
|
| rs780711806 | 206 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs748583369 | 206 | Y>F | No |
ExAC gnomAD |
|
| rs2066753526 | 208 | A>V | No | Ensembl | |
| rs751009983 | 209 | G>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 210 | Y>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066752897 | 211 | L>V | No | gnomAD | |
| rs1367523730 | 213 | T>A | No | gnomAD | |
| rs1367523730 | 213 | T>P | No | gnomAD | |
| rs376374684 | 214 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 215 | T>S | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066751793 | 216 | Q>R | No | Ensembl | |
| rs921924761 | 217 | Q>L | No | TOPMed | |
| TCGA novel | 218 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775396375 | 219 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs75911819 | 220 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2066751196 | 220 | A>T | No | TOPMed | |
| rs75911819 | 220 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs774293656 | 222 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs771239742 | 223 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs763024214 | 224 | D>N | No |
ExAC gnomAD |
|
| rs376560284 | 225 | G>D | No |
ESP ExAC gnomAD |
|
| rs770104035 | 225 | G>S | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 225 | G>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs375502604 | 226 | P>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs375502604 | 226 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1211334712 | 227 | Y>S | No |
TOPMed gnomAD |
|
| rs746589854 | 228 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs374549222 | 229 | F>V | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 230 | F>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1255737072 | 231 | V>A | No | TOPMed | |
| rs1191748765 | 231 | V>L | No | gnomAD | |
| rs758074370 | 232 | F>S | No |
ExAC TOPMed gnomAD |
|
| COSM4105104 | 232 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2093443075 | 233 | N>I | No | gnomAD | |
| rs778877074 | 236 | D>H | No |
ExAC gnomAD |
|
| rs2066747172 | 237 | K>E | No | gnomAD | |
| rs753658408 | 237 | K>R | No |
ExAC gnomAD |
|
| rs764196872 | 239 | P>L | No |
ExAC gnomAD |
|
| rs1454388486 | 239 | P>S | No | gnomAD | |
| rs1450418662 | 240 | A>S | No |
TOPMed gnomAD |
|
| rs201878905 | 241 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs766298920 | 241 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs773373233 | 243 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1227754334 | 243 | R>H | No |
TOPMed gnomAD |
|
| rs372225867 | 244 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1292732087 | 245 | L>M | No | Ensembl | |
| rs779647430 | 248 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs910158916 | 248 | R>H | No | gnomAD | |
| rs985664639 | 249 | M>I | No | gnomAD | |
| rs2066743127 | 249 | M>V | No | Ensembl | |
| rs771435200 | 250 | C>Y | No |
ExAC gnomAD |
|
| rs28561594 | 252 | E>G | No |
ESP ExAC TOPMed |
|
| rs1601728564 | 253 | D>A | No | Ensembl | |
| TCGA novel | 253 | D>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 253 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1255644373 | 254 | P>L | No | Ensembl | |
| rs1209152483 | 254 | P>T | No |
TOPMed gnomAD |
|
| rs1486280033 | 255 | N>D | No | TOPMed | |
| rs1207006502 | 258 | S>A | No | TOPMed | |
| rs577027165 | 260 | L>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1324027168 | 262 | M>I | No | gnomAD | |
| rs2066740947 | 262 | M>V | No | TOPMed | |
| rs1601728521 | 263 | D>A | No | Ensembl | |
| rs1377601683 | 263 | D>N | No |
TOPMed gnomAD |
|
| rs2066740036 | 265 | Q>E | No | Ensembl | |
|
COSM3555705 rs200109107 |
265 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC TOPMed gnomAD NCI-TCGA Cosmic |
| COSM1035417 | 266 | C>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs756079656 | 267 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs756079656 | 267 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs777651523 | 267 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs374601429 | 268 | D>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs374601429 | 268 | D>G | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 268 | D>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374601429 | 268 | D>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs1569174092 | 269 | P>A | No | Ensembl | |
| rs1569174092 | 269 | P>S | No | Ensembl | |
| rs1020040133 | 270 | D>A | No | Ensembl | |
| rs750469859 | 270 | D>N | No |
ExAC gnomAD |
|
| rs750469859 | 270 | D>Y | No |
ExAC gnomAD |
|
| rs765508447 | 271 | I>F | No |
ExAC TOPMed gnomAD |
|
| rs765508447 | 271 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs28523652 | 272 | H>P | No | Ensembl | |
| rs886582997 | 272 | H>Q | No |
TOPMed gnomAD |
|
| rs761966193 | 273 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs377609672 | 274 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs761267538 | 276 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs374654956 | 278 | T>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs374654956 | 278 | T>P | No |
ESP ExAC TOPMed gnomAD |
|
| rs745439137 | 280 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs1159582667 | 281 | A>G | No | gnomAD | |
| rs773988692 | 281 | A>S | No |
ExAC gnomAD |
|
| rs200815418 | 282 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM4105103 | 282 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs747951490 | 284 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1191011058 | 285 | A>S | No | TOPMed | |
|
rs554117465 COSM3842937 |
286 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs1441846538 | 287 | P>S | No | gnomAD | |
| rs2066734788 | 289 | S>C | No | TOPMed | |
| rs1206774356 | 290 | G>D | No | gnomAD | |
| rs571462806 | 290 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1569173910 | 291 | R>K | No | Ensembl | |
| rs1356955662 | 296 | V>L | No | gnomAD | |
| rs1468212585 | 298 | S>C | No | Ensembl | |
| rs775923722 | 300 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs2066732977 | 300 | D>V | No | TOPMed | |
| rs1435614645 | 301 | S>N | No |
TOPMed gnomAD |
|
| rs1435614645 | 301 | S>T | No |
TOPMed gnomAD |
|
| rs760065938 | 302 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs767968899 | 302 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs770522658 | 303 | S>I | No |
ExAC gnomAD |
|
| rs769456290 | 305 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs769456290 | 305 | G>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 306 | G>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1233981945 | 307 | P>L | No |
TOPMed gnomAD |
|
| TCGA novel | 308 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1180279837 | 308 | G>S | No | gnomAD | |
| TCGA novel | 308 | G>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066730528 | 308 | G>V | No | Ensembl | |
| rs1242732256 | 309 | A>S | No |
TOPMed gnomAD |
|
| rs1242732256 | 309 | A>T | No |
TOPMed gnomAD |
|
| rs754877775 | 309 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs371292339 | 310 | G>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs772771317 | 313 | L>P | No |
TOPMed gnomAD |
|
| rs764294104 | 314 | F>Y | No |
ExAC gnomAD |
|
| rs756348985 | 315 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs369038951 | 315 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP NCI-TCGA TOPMed gnomAD |
| rs767853611 | 317 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1601728158 | 317 | D>Y | No | Ensembl | |
|
rs28379706 VAR_050600 |
318 | K>E | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs1442300968 | 319 | V>M | No | gnomAD | |
| rs371959367 | 320 | H>L | No |
ESP ExAC gnomAD |
|
| rs375153131 | 321 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs769389095 | 326 | N>K | No |
ExAC gnomAD |
|
| rs1447810081 | 327 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs776323520 | 327 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs776323520 | 327 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs746784728 | 328 | N>K | No |
ExAC gnomAD |
|
| rs1220605772 | 329 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs780008242 | 331 | Y>* | No |
ExAC gnomAD |
|
| rs201390815 | 332 | T>I | No | Ensembl | |
| TCGA novel | 332 | T>K | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs112883077 | 334 | T>P | No | Ensembl | |
| rs749432150 | 335 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs771996791 | 335 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1303532696 | 336 | E>K | No |
TOPMed gnomAD |
|
| COSM726728 | 337 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372336824 | 338 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1417080 rs530051710 |
338 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs530051710 | 338 | R>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs372336824 | 338 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs752921231 | 339 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs1601728010 | 340 | I>N | No | Ensembl | |
| rs755439325 | 340 | I>V | No |
ExAC gnomAD |
|
| rs1169888392 | 343 | K>E | No | gnomAD | |
|
COSM6018313 rs559951398 |
344 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs2066722204 | 345 | F>L | No | TOPMed | |
| rs1235380008 | 346 | H>L | No | TOPMed | |
| rs1389583451 | 346 | H>Y | No | gnomAD | |
| rs763538802 | 347 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs764764946 | 348 | D>G | No |
ExAC gnomAD |
|
| rs2066721288 | 348 | D>N | No |
TOPMed gnomAD |
|
| rs1601727941 | 349 | I>T | No | Ensembl | |
| rs761396649 | 350 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs760534896 | 352 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs760534896 | 352 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1020073040 | 353 | G>S | No |
TOPMed gnomAD |
|
| rs745754194 | 354 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs532708125 | 355 | A>E | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs777742714 | 355 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs532708125 | 355 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs755245449 | 356 | P>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs565122764 | 356 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2147517945 | 357 | G>S | No | Ensembl | |
| rs1601725717 | 360 | K>N | No | Ensembl | |
| COSM1417079 | 361 | S>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768815439 | 361 | S>R | No |
ExAC gnomAD |
|
| COSM3800287 | 362 | F>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1449257751 | 364 | C>R | No | TOPMed | |
| rs1569172847 | 366 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs1601725692 | 368 | H>P | No | Ensembl | |
| rs1464189618 | 370 | P>L | No | Ensembl | |
| rs779092516 | 372 | P>L | No |
ExAC gnomAD |
|
| rs2066681903 | 373 | L>P | No | TOPMed | |
| rs777290861 | 374 | G>S | No |
ExAC gnomAD |
|
| rs370637750 | 376 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs143051588 | 376 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs766018907 | 377 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 377 | D>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1384922122 | 378 | G>R | No |
TOPMed gnomAD |
|
| rs750436164 | 379 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs750436164 | 379 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs2147517192 | 379 | L>P | No | Ensembl | |
| rs2066679979 | 380 | R>I | No | TOPMed | |
| rs1368115644 | 381 | G>S | No |
TOPMed gnomAD |
|
| rs760906220 | 382 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs772355652 | 383 | A>P | No |
ExAC gnomAD |
|
| rs573754705 | 384 | V>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs573754705 | 384 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs770278125 | 386 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs747712044 | 387 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs747712044 | 387 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs754662011 | 387 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs754662011 | 387 | R>P | No |
ExAC TOPMed gnomAD |
|
| rs751131363 | 388 | G>V | No |
ExAC gnomAD |
|
| rs758310101 | 393 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs758310101 | 393 | T>R | No |
ExAC TOPMed gnomAD |
|
| COSM5549755 | 394 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs767458963 | 395 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs767458963 | 395 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs376982545 | 396 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1174766050 | 398 | A>T | No |
TOPMed gnomAD |
|
| rs547358862 | 399 | A>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs547358862 | 399 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1302596108 | 400 | E>D | No |
TOPMed gnomAD |
|
|
COSM1035416 rs776234564 |
400 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs372964527 | 401 | N>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs28591681 | 401 | N>S | No | Ensembl | |
| rs1181547336 | 404 | T>A | No | gnomAD | |
| rs746497917 | 404 | T>I | No |
ExAC gnomAD |
|
| TCGA novel | 411 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778786971 | 412 | D>H | No |
ExAC gnomAD |
|
| rs778786971 | 412 | D>N | No |
ExAC gnomAD |
|
| rs2066671814 | 413 | G>S | No | Ensembl | |
| rs145844372 | 414 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs757294277 | 414 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2066670990 | 416 | L>F | No | Ensembl | |
| rs1009594300 | 418 | V>A | No | Ensembl | |
| rs1354097895 | 418 | V>L | No |
TOPMed gnomAD |
|
| rs375132799 | 419 | Y>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs775174241 | 420 | L>I | No |
ExAC TOPMed gnomAD |
|
| rs775174241 | 420 | L>V | No |
ExAC TOPMed gnomAD |
|
| rs147728569 | 421 | T>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2066660744 | 421 | T>S | No | gnomAD | |
| rs1298989097 | 422 | P>A | No | TOPMed | |
| rs1416300554 | 422 | P>L | No |
TOPMed gnomAD |
|
| rs1416300554 | 422 | P>Q | No |
TOPMed gnomAD |
|
| rs770698601 | 423 | D>E | No |
ExAC gnomAD |
|
| rs2066659395 | 423 | D>G | No | gnomAD | |
| rs544468493 | 424 | G>D | No | gnomAD | |
| rs1390877190 | 424 | G>S | No | gnomAD | |
| rs544468493 | 424 | G>V | No | gnomAD | |
| rs777563259 | 425 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs1005391338 | 426 | S>F | No |
TOPMed gnomAD |
|
| rs1208878874 | 428 | E>G | No | gnomAD | |
| rs1231209117 | 430 | D>G | No | gnomAD | |
| rs1299203677 | 430 | D>N | No |
TOPMed gnomAD |
|
| COSM3371955 | 431 | S>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs750470230 | 432 | I>V | No |
ExAC TOPMed gnomAD |
|
| COSM3555703 | 433 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1269962777 | 434 | V>L | No | gnomAD | |
| TCGA novel | 434 | V>Y | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1437710944 | 435 | E>A | No | gnomAD | |
| rs2066656219 | 435 | E>D | No |
TOPMed gnomAD |
|
| rs905405369 | 438 | K>E | No | Ensembl | |
| rs1045647927 | 438 | K>N | No | Ensembl | |
| rs1351989403 | 438 | K>R | No |
TOPMed gnomAD |
|
| rs374592428 | 441 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs374592428 | 441 | K>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1371194693 | 441 | K>R | No |
TOPMed gnomAD |
|
| rs1371194693 | 441 | K>T | No |
TOPMed gnomAD |
|
| rs1170606000 | 442 | R>C | No |
TOPMed gnomAD |
|
| rs754158572 | 442 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2066653935 | 443 | D>G | No | TOPMed | |
| rs764481494 | 443 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs2066653647 | 445 | V>L | No | Ensembl | |
| rs761110627 | 447 | S>C | No |
ExAC gnomAD |
|
| rs1056791152 | 447 | S>P | No | Ensembl | |
| rs376295504 | 448 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1483197768 | 449 | D>Y | No | gnomAD | |
| rs2066652423 | 450 | L>V | No | TOPMed | |
| rs759187654 | 451 | G>A | No |
ExAC gnomAD |
|
| rs759187654 | 451 | G>V | No |
ExAC gnomAD |
|
| rs1202484286 | 452 | S>I | No | gnomAD | |
| rs1202484286 | 452 | S>N | No | gnomAD | |
| rs372467611 | 452 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs1346124430 | 453 | L>V | No |
TOPMed gnomAD |
|
| COSM6095526 | 454 | Y>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772909474 | 455 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 455 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1417078 | 456 | M>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1404533598 | 458 | Q>* | No | gnomAD | |
| rs747914479 | 459 | D>N | No |
ExAC gnomAD |
|
| rs2066650013 | 460 | K>R | No | Ensembl | |
| rs1226756894 | 461 | V>A | No | gnomAD | |
| TCGA novel | 461 | V>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752061694 | 462 | F>L | No |
ExAC gnomAD |
|
| rs192965378 | 463 | R>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs988685059 | 463 | R>W | No |
TOPMed gnomAD |
|
| rs370042051 | 465 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs370042051 | 465 | P>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1174947935 | 466 | V>M | No |
TOPMed gnomAD |
|
| rs935182746 | 468 | E>K | No | Ensembl | |
| rs2066592364 | 469 | C>R | No | TOPMed | |
| rs1410165755 | 471 | S>N | No | gnomAD | |
| rs763829959 | 473 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs763829959 | 473 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs2066591285 | 474 | T>I | No | TOPMed | |
| rs2066591285 | 474 | T>N | No | TOPMed | |
| rs2147500956 | 474 | T>P | No | Ensembl | |
| rs2147500905 | 475 | C>S | No | Ensembl | |
| rs1298731036 | 477 | Q>E | No |
TOPMed gnomAD |
|
| rs1298731036 | 477 | Q>K | No |
TOPMed gnomAD |
|
| rs775541473 | 477 | Q>R | No |
ExAC gnomAD |
|
| rs1450674690 | 478 | C>Y | No | gnomAD | |
| rs770096187 | 479 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs770096187 | 479 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs773103653 | 479 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1266189940 | 480 | D>N | No |
TOPMed gnomAD |
|
| COSM1035415 | 481 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1224065571 | 481 | S>T | No | gnomAD | |
| rs866516626 | 482 | Q>K | No | Ensembl | |
| rs1244295436 | 482 | Q>R | No |
TOPMed gnomAD |
|
| rs781705361 | 486 | C>Y | No |
ExAC gnomAD |
|
| TCGA novel | 487 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs747304123 | 489 | C>G | No |
ExAC gnomAD |
|
| rs749954299 | 490 | V>A | No |
ExAC gnomAD |
|
| rs758817773 | 490 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs758817773 | 490 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1601721271 | 491 | V>A | No | Ensembl | |
| rs1364368096 | 491 | V>F | No |
TOPMed gnomAD |
|
| rs1364368096 | 491 | V>I | No |
TOPMed gnomAD |
|
| rs1419000468 | 492 | E>A | No |
TOPMed gnomAD |
|
| TCGA novel | 492 | E>D | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs763707512 | 492 | E>K | No |
ExAC gnomAD |
|
| rs763707512 | 492 | E>Q | No |
ExAC gnomAD |
|
| rs1191484055 | 494 | R>Q | No |
TOPMed gnomAD |
|
| rs1204353756 | 495 | C>F | No |
TOPMed gnomAD |
|
| rs113752649 | 495 | C>G | No | Ensembl | |
| rs1012489724 | 496 | T>A | No | Ensembl | |
| rs371186063 | 497 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779460198 | 497 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs371186063 | 497 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs771412072 | 498 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs777192973 | 500 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs777192973 | 500 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs1258262069 | 501 | C>Y | No | gnomAD | |
| rs752246192 | 502 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs752246192 | 502 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs755826651 | 502 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs751397463 | 503 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs754771727 | 503 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs578011591 | 504 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs558651178 COSM5628653 |
505 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 506 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs760785944 | 507 | A>P | No |
ExAC gnomAD |
|
| rs1255977405 | 507 | A>V | No |
TOPMed gnomAD |
|
| rs1321695799 | 508 | S>T | No |
TOPMed gnomAD |
|
| rs1459003711 | 509 | H>Q | No | gnomAD | |
| rs2066566803 | 509 | H>R | No | TOPMed | |
| rs374467610 | 514 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs774637101 | 516 | K>R | No |
ExAC gnomAD |
|
| rs573977985 | 517 | S>F | No |
1000Genomes ExAC gnomAD |
|
| rs2066565411 | 518 | C>Y | No | Ensembl | |
| rs370179188 | 519 | V>M | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1444914291 | 520 | A>S | No | gnomAD | |
| rs930904680 | 521 | V>I | No |
1000Genomes TOPMed gnomAD |
|
| rs2066564239 | 522 | T>A | No | Ensembl | |
| TCGA novel | 522 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs375580670 | 524 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs746814341 | 527 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs920837059 | 528 | N>D | No | Ensembl | |
| rs758431542 | 529 | M>I | No |
ExAC gnomAD |
|
| rs965224973 | 529 | M>L | No | Ensembl | |
| rs2066562655 | 530 | S>T | No | TOPMed | |
| rs1325057484 | 531 | R>Q | No | gnomAD | |
| rs1433414091 | 531 | R>W | No |
TOPMed gnomAD |
|
| rs866654904 | 532 | R>W | No |
TOPMed gnomAD |
|
| rs1406198220 | 533 | A>T | No |
TOPMed gnomAD |
|
| rs2147495501 | 534 | Q>P | No | Ensembl | |
| rs1384857397 | 535 | G>E | No | TOPMed | |
| rs746118447 | 535 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs746118447 | 535 | G>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 536 | E>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2147495358 | 536 | E>G | No | Ensembl | |
| rs1450215526 | 536 | E>K | No | TOPMed | |
| rs2147488113 | 538 | Q>H | No | Ensembl | |
| rs2066524719 | 538 | Q>R | No | TOPMed | |
| rs923713459 | 541 | V>A | No | Ensembl | |
| rs768199498 | 541 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs768199498 | 541 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs746620222 | 542 | S>G | No |
ExAC TOPMed gnomAD |
|
| rs1697709939 | 542 | S>R | No | TOPMed | |
| rs1353922189 | 544 | L>F | No | gnomAD | |
| rs374129632 | 544 | L>R | No |
ESP TOPMed gnomAD |
|
| rs1470797614 | 545 | P>L | No | gnomAD | |
| rs2066522892 | 546 | A>V | No | Ensembl | |
| rs758308131 | 549 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1472733540 | 550 | E>K | No |
TOPMed gnomAD |
|
| rs370325511 | 551 | D>E | No |
1000Genomes gnomAD |
|
| rs745652433 | 551 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs574088314 | 551 | D>V | No |
1000Genomes ExAC gnomAD |
|
| rs376816438 | 552 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs376816438 | 552 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1013266469 | 556 | L>H | No | TOPMed | |
| rs2066520910 | 556 | L>V | No | Ensembl | |
| rs80161851 | 557 | F>L | No | Ensembl | |
| rs781287988 | 557 | F>S | No |
ExAC TOPMed gnomAD |
|
| rs781287988 | 557 | F>Y | No |
ExAC TOPMed gnomAD |
|
| rs2066519965 | 559 | E>D | No | TOPMed | |
| rs1223098951 | 559 | E>G | No |
TOPMed gnomAD |
|
| rs2066519818 | 560 | S>L | No | Ensembl | |
| rs1676455744 | 560 | S>T | No | gnomAD | |
| TCGA novel | 560 | S>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1309222896 | 561 | P>L | No |
TOPMed gnomAD |
|
| rs1372707455 | 562 | P>S | No |
TOPMed gnomAD |
|
| rs1372707455 | 562 | P>T | No |
TOPMed gnomAD |
|
| rs1601717965 | 563 | H>P | No | Ensembl | |
| rs1284654575 | 563 | H>Y | No |
TOPMed gnomAD |
|
| rs1476539274 | 564 | P>S | No |
TOPMed gnomAD |
|
| rs1476539274 | 564 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs750788711 | 565 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs750788711 | 565 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs765677296 | 566 | R>C | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs762315379 | 566 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs765677296 | 566 | R>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1156496011 | 567 | V>L | No |
TOPMed gnomAD |
|
| rs1156496011 | 567 | V>M | No |
TOPMed gnomAD |
|
| rs1601717911 | 568 | E>G | No | Ensembl | |
| rs2147486561 | 568 | E>K | No | Ensembl | |
| rs539598184 | 569 | G>C | No |
1000Genomes TOPMed gnomAD |
|
| rs539598184 | 569 | G>S | No |
1000Genomes TOPMed gnomAD |
|
| rs1447016564 | 570 | E>K | No |
TOPMed gnomAD |
|
| rs994545360 | 571 | A>T | No |
TOPMed gnomAD |
|
| rs2066516462 | 571 | A>V | No | gnomAD | |
| rs768260684 | 572 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1036427656 | 573 | I>M | No |
TOPMed gnomAD |
|
| rs1262645431 | 573 | I>V | No | gnomAD | |
| TCGA novel | 574 | C>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1316368705 | 575 | N>H | No |
TOPMed gnomAD |
|
| rs1601717866 | 575 | N>T | No | Ensembl | |
| rs760231402 | 577 | P>R | No |
ExAC TOPMed gnomAD |
|
| rs754908392 | 578 | S>G | No | Ensembl | |
| rs1281340664 | 578 | S>N | No | Ensembl | |
| rs1569169738 | 579 | S>N | No | Ensembl | |
| rs771834961 | 582 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs1363825688 | 583 | T>K | No | gnomAD | |
| rs2066513152 | 584 | P>A | No | Ensembl | |
| rs557311559 | 584 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs890732555 | 585 | P>L | No |
TOPMed gnomAD |
|
| rs890732555 | 585 | P>R | No |
TOPMed gnomAD |
|
| rs749196497 | 585 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs777552616 | 586 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs777552616 | 586 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs1429426396 | 587 | Q>H | No |
TOPMed gnomAD |
|
| rs755094925 | 587 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs755094925 | 587 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs201989267 | 589 | H>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs775569687 | 590 | V>M | No |
ExAC gnomAD |
|
| rs1248248221 | 592 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs745931051 | 593 | T>I | No |
ExAC gnomAD |
|
| rs779253388 | 594 | I>F | No | ExAC | |
| rs11547733 | 594 | I>M | No | Ensembl | |
| rs1264811027 | 594 | I>S | No |
TOPMed gnomAD |
|
| rs779253388 | 594 | I>V | No | ExAC | |
| rs757361795 | 595 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs778239432 | 598 | L>F | No |
ExAC TOPMed gnomAD |
|
|
rs1333488993 COSM3065379 |
600 | R>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1333488993 | 600 | R>G | No |
TOPMed gnomAD |
|
| rs756708347 | 600 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs2066448057 | 601 | G>S | No | Ensembl | |
| rs1407258647 | 602 | N>D | No |
1000Genomes gnomAD |
|
| rs1339160904 | 604 | F>V | No | gnomAD | |
| rs148722170 | 605 | L>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs751022390 | 606 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2066446476 | 609 | Q>R | No | Ensembl | |
| rs2066446041 | 611 | P>R | No | TOPMed | |
| rs200418993 | 611 | P>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2066445908 | 612 | F>S | No |
TOPMed gnomAD |
|
| rs761575337 | 614 | D>N | No |
ExAC gnomAD |
|
| rs371223423 | 616 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs745972033 | 616 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1601713769 | 617 | Q>P | No | Ensembl | |
| rs1206548459 | 618 | A>V | No | gnomAD | |
| rs565870583 | 619 | M>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2147470624 | 619 | M>R | No | Ensembl | |
| rs1421329356 | 621 | L>Q | No |
TOPMed gnomAD |
|
| rs961858625 | 621 | L>V | No | Ensembl | |
| rs1384258254 | 622 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1317605470 | 623 | E>K | No | gnomAD | |
| rs1340264712 | 624 | N>K | No | gnomAD | |
| rs781106994 | 624 | N>S | No |
ExAC gnomAD |
|
| rs781106994 | 624 | N>T | No |
ExAC gnomAD |
|
| rs1402694378 | 626 | P>L | No | gnomAD | |
| rs747427027 | 628 | I>M | No |
ExAC gnomAD |
|
| COSM3555701 | 629 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1161549412 | 631 | V>L | No |
TOPMed gnomAD |
|
| rs1161549412 | 631 | V>M | No |
TOPMed gnomAD |
|
| rs749920297 | 632 | S>N | No |
ExAC TOPMed gnomAD |
|
| rs749920297 | 632 | S>T | No |
ExAC TOPMed gnomAD |
|
| rs778550298 | 633 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs778550298 | 633 | N>T | No |
ExAC TOPMed gnomAD |
|
| rs756834081 | 634 | R>C | No |
ExAC gnomAD |
|
| rs144108995 | 634 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs144108995 | 634 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1199714162 | 635 | W>* | No | gnomAD | |
| rs1199714162 | 635 | W>C | No | gnomAD | |
| rs1601713327 | 635 | W>G | No | Ensembl | |
| rs199698162 | 636 | T>P | No |
ExAC gnomAD |
|
| rs1045681680 | 636 | T>S | No | TOPMed | |
| rs2066432925 | 639 | W>* | No | TOPMed | |
| rs1445136737 | 640 | D>E | No | gnomAD | |
| rs2066432430 | 641 | L>P | No | TOPMed | |
| rs2066432586 | 641 | L>V | No | Ensembl | |
| rs752612256 | 642 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs767483052 | 642 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs190686647 | 644 | H>Q | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2066431676 | 644 | H>R | No | Ensembl | |
| rs2147468241 | 645 | E>* | No | Ensembl | |
| rs1347435068 | 645 | E>D | No | gnomAD | |
| rs369676765 | 647 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs371571515 | 647 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 648 | E>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM6095527 | 650 | S>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1004979336 | 650 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs530363071 | 651 | P>L | No |
1000Genomes TOPMed gnomAD |
|
| rs2066430031 | 653 | P>R | No | TOPMed | |
| rs375149704 | 653 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1269324664 | 655 | D>N | No | Ensembl | |
| rs1176657220 | 656 | G>S | No | gnomAD | |
| rs1197970133 | 656 | G>V | No |
TOPMed gnomAD |
|
| rs2066429338 | 657 | I>V | No |
TOPMed gnomAD |
|
| rs745385290 | 658 | V>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs201060198 | 658 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1477987303 | 659 | R>C | No |
TOPMed gnomAD |
|
| rs778354203 | 659 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs2066428372 | 660 | A>V | No | gnomAD | |
| rs756924022 | 661 | H>Y | No |
ExAC gnomAD |
|
| rs1481801278 | 662 | M>T | No | gnomAD | |
| rs748937712 | 662 | M>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2066419584 | 663 | E>K | No |
TOPMed gnomAD |
|
| rs1301853661 | 664 | D>E | No |
TOPMed gnomAD |
|
| rs751596451 | 664 | D>N | No |
ExAC gnomAD |
|
| rs138005986 | 665 | S>C | No |
1000Genomes ExAC gnomAD |
|
| rs138005986 | 665 | S>G | No |
1000Genomes ExAC gnomAD |
|
| rs1346427963 | 665 | S>I | No | gnomAD | |
| rs2066418568 | 667 | P>T | No | TOPMed | |
| COSM4849340 | 668 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1156385285 | 668 | Q>H | No | gnomAD | |
| rs1010102713 | 670 | L>R | No |
TOPMed gnomAD |
|
| rs1164785055 | 671 | G>R | No |
TOPMed gnomAD |
|
| rs1196476125 | 673 | S>G | No |
TOPMed gnomAD |
|
| rs267606288 | 676 | V>G | No |
TOPMed gnomAD |
|
| rs753025129 | 677 | I>F | No |
ExAC gnomAD |
|
| COSM3555700 | 678 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs774901026 | 679 | M>I | No |
ExAC TOPMed gnomAD |
|
| rs759858793 | 679 | M>L | No |
ExAC TOPMed gnomAD |
|
| rs1292339447 | 680 | N>S | No | gnomAD | |
| rs772714834 | 681 | H>Q | No |
ExAC TOPMed gnomAD |
|
| rs747791337 | 682 | E>A | No |
ExAC gnomAD |
|
| TCGA novel | 682 | E>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769364893 | 682 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs780970195 | 683 | T>A | No |
ExAC gnomAD |
|
| rs768612857 | 683 | T>R | No |
ExAC gnomAD |
|
| TCGA novel | 684 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 685 | V>D | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs780199079 | 686 | N>K | No |
ExAC TOPMed gnomAD |
|
| rs758286991 | 688 | Q>E | No |
ExAC gnomAD |
|
| rs2066414991 | 689 | G>S | No | TOPMed | |
| rs2066414848 | 690 | K>R | No | gnomAD | |
| rs1304165628 | 693 | D>H | No | TOPMed | |
| rs777859721 | 693 | D>V | No |
ExAC gnomAD |
|
| rs375588549 | 695 | V>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1569167906 | 696 | K>E | No | Ensembl | |
| rs200360490 | 697 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs2066287681 | 697 | G>V | No | Ensembl | |
| rs1266767480 | 698 | S>F | No | TOPMed | |
| rs2066287421 | 698 | S>P | No | Ensembl | |
| rs1184631640 | 699 | S>F | No |
TOPMed gnomAD |
|
| rs759068611 | 699 | S>T | No |
ExAC gnomAD |
|
| rs766214363 | 701 | H>R | No |
ExAC TOPMed gnomAD |
|
| rs769706219 | 702 | V>A | No |
ExAC gnomAD |
|
| rs773204897 | 702 | V>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 703 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066285665 | 704 | S>N | No | Ensembl | |
| rs200637738 | 705 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| COSM1035414 | 705 | D>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs772372051 | 706 | L>F | No |
ExAC gnomAD |
|
| rs1356074125 | 707 | L>F | No |
TOPMed gnomAD |
|
| rs746123380 | 707 | L>P | No |
ExAC TOPMed gnomAD |
|
| rs746123380 | 707 | L>R | No |
ExAC TOPMed gnomAD |
|
| rs1356074125 | 707 | L>V | No |
TOPMed gnomAD |
|
| rs553785965 | 709 | F>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778433550 | 710 | M>R | No |
ExAC gnomAD |
|
| rs778433550 | 710 | M>T | No |
ExAC gnomAD |
|
| rs1405492879 | 710 | M>V | No | gnomAD | |
| rs376746165 | 712 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs752279615 | 714 | T>I | No |
ExAC gnomAD |
|
| rs1176769334 | 716 | Q>R | No | gnomAD | |
| rs780516842 | 718 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs780516842 | 718 | S>F | No |
ExAC TOPMed gnomAD |
|
| rs2066283184 | 719 | G>R | No | TOPMed | |
| rs1476609267 | 720 | T>A | No | gnomAD | |
| TCGA novel | 720 | T>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374900808 | 722 | A>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs374900808 | 722 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs1481129048 | 722 | A>V | No | gnomAD | |
|
COSM1190495 rs762799219 |
724 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
|
rs201521733 COSM3786401 |
724 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs750161648 | 726 | P>R | No |
ExAC gnomAD |
|
| rs2066281853 | 727 | K>E | No | TOPMed | |
| rs1274520275 | 729 | S>F | No | TOPMed | |
| rs2066247278 | 730 | H>P | No | Ensembl | |
| rs769168176 | 731 | D>N | No | gnomAD | |
| TCGA novel | 733 | N>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs200165560 | 735 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs372542578 | 736 | L>M | No |
ESP TOPMed gnomAD |
|
| rs777066522 | 737 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs2066246072 | 737 | P>S | No | TOPMed | |
| rs746527136 | 740 | L>F | No |
ExAC gnomAD |
|
| rs370061884 | 742 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs778621151 | 743 | K>M | No |
ExAC TOPMed gnomAD |
|
| rs757056511 | 744 | S>F | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs1749587286 |
745 | Y>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| TCGA novel | 745 | Y>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs532944576 COSM1035413 |
746 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs766613051 | 748 | N>S | No |
ExAC gnomAD |
|
| rs1478212254 | 749 | I>V | No |
TOPMed gnomAD |
|
| rs1214386254 | 750 | D>E | No | gnomAD | |
| rs1332617247 | 750 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs867846722 | 754 | H>R | No |
TOPMed gnomAD |
|
| rs773637239 | 754 | H>Y | No |
ExAC gnomAD |
|
| rs761021468 | 756 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs761021468 | 756 | T>N | No |
ExAC TOPMed gnomAD |
|
| rs1438903009 | 757 | L>V | No |
TOPMed gnomAD |
|
| rs2147424977 | 758 | Y>C | No | Ensembl | |
| rs79966207 | 759 | N>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772506820 | 759 | N>S | No |
ExAC gnomAD |
|
| rs1304594338 | 760 | C>Y | No | gnomAD | |
| rs1464607448 | 763 | G>D | No | gnomAD | |
| rs1159157606 | 764 | R>C | No | gnomAD | |
|
COSM1417072 rs1441358342 |
764 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1441358342 | 764 | R>L | No |
TOPMed gnomAD |
|
| rs1159157606 | 764 | R>S | No | gnomAD | |
| rs538514666 | 765 | S>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
COSM1035412 rs1442577954 |
766 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs749018022 | 769 | L>M | No |
ExAC gnomAD |
|
| rs1456049111 | 770 | C>Y | No | TOPMed | |
| rs1280294689 | 771 | R>L | No |
TOPMed gnomAD |
|
| rs1280294689 | 771 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs777258572 | 771 | R>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 771 | R>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs781292997 | 773 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1238613070 | 774 | N>D | No |
TOPMed gnomAD |
|
| rs1238613070 | 774 | N>H | No |
TOPMed gnomAD |
|
| rs1341368423 | 775 | P>L | No | gnomAD | |
| rs374234427 | 776 | D>N | No |
ESP ExAC TOPMed gnomAD |
|
| rs374234427 | 776 | D>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1421388101 | 779 | C>Y | No | gnomAD | |
| rs534402671 | 780 | A>T | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1157753906 | 780 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1369893535 | 781 | W>* | No | gnomAD | |
| TCGA novel | 781 | W>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752989186 | 782 | C>Y | No |
ExAC gnomAD |
|
| rs551456267 | 783 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs774697614 | 784 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs2066213863 | 785 | Q>H | No |
TOPMed gnomAD |
|
| rs770513867 | 785 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs770513867 | 785 | Q>R | No |
ExAC TOPMed gnomAD |
|
| rs1204976368 | 786 | S>N | No | gnomAD | |
| rs1204976368 | 786 | S>T | No | gnomAD | |
| rs762363550 | 787 | R>G | No |
ExAC TOPMed gnomAD |
|
| rs2066213329 | 787 | R>S | No | Ensembl | |
| rs762363550 | 787 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs2066213180 | 788 | C>Y | No | TOPMed | |
| rs769514548 | 789 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs769514548 | 789 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs374465427 | 790 | Y>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs768741557 | 791 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs1392574312 | 792 | A>D | No | gnomAD | |
| rs747019847 | 792 | A>T | No |
ExAC gnomAD |
|
| rs2066211377 | 795 | N>D | No | TOPMed | |
| rs757427704 | 796 | T>I | No |
ExAC gnomAD |
|
|
COSM4105100 rs1476547887 |
797 | T>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1261718526 | 797 | T>I | No | gnomAD | |
| rs953636958 | 799 | E>* | No |
TOPMed gnomAD |
|
| rs953636958 | 799 | E>K | No |
TOPMed gnomAD |
|
| rs756284707 | 800 | C>* | No | ExAC | |
| TCGA novel | 801 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1208299039 | 801 | P>Q | No | gnomAD | |
| rs375572338 | 802 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1258944875 | 802 | P>S | No | gnomAD | |
| TCGA novel | 802 | P>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066209118 | 803 | P>H | No |
TOPMed gnomAD |
|
| rs370412897 | 804 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs370412897 | 804 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1212157084 | 805 | I>V | No | TOPMed | |
| rs1332625528 | 806 | T>S | No | gnomAD | |
| rs2066207967 | 807 | R>K | No | TOPMed | |
| rs763643132 | 808 | I>M | No |
ExAC gnomAD |
|
| rs200426394 | 809 | Q>L | No |
ExAC TOPMed gnomAD |
|
| rs28470336 | 812 | T>A | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs772065446 | 812 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs28470336 | 812 | T>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs774241133 | 814 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs774241133 | 814 | P>T | No |
ExAC TOPMed gnomAD |
|
| rs965072605 | 815 | L>P | No | Ensembl | |
| rs2066197781 | 816 | G>A | No | Ensembl | |
| rs368997252 | 816 | G>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs368997252 | 816 | G>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1372240792 | 817 | G>R | No | gnomAD | |
| rs562878563 | 818 | G>S | No | 1000Genomes | |
| rs781408320 | 819 | I>V | No |
ExAC gnomAD |
|
| rs376548374 | 820 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs747432493 | 820 | R>H | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs747432493 | 820 | R>P | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs780232837 | 821 | I>L | No |
ExAC TOPMed gnomAD |
|
| rs758772028 | 822 | T>A | No |
ExAC gnomAD |
|
| rs751011693 | 822 | T>I | No |
ExAC gnomAD |
|
| rs751011693 | 822 | T>S | No |
ExAC gnomAD |
|
| rs11547731 | 823 | I>F | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2066195066 | 823 | I>T | No |
TOPMed gnomAD |
|
|
VAR_061537 rs11547731 |
823 | I>V | No |
UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs2066194014 | 826 | S>F | No | TOPMed | |
| rs534546225 | 827 | N>D | No |
1000Genomes ExAC TOPMed gnomAD |
|
| COSM1035411 | 827 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 827 | N>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1308291 | 828 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1289387729 | 829 | G>S | No |
TOPMed gnomAD |
|
| rs376293364 | 830 | V>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs376293364 | 830 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs770795215 | 833 | G>E | No |
ExAC TOPMed gnomAD |
|
| rs770795215 | 833 | G>V | No |
ExAC TOPMed gnomAD |
|
| rs776534685 | 836 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs376030784 | 837 | R>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs2066191856 | 839 | S>F | No | gnomAD | |
| rs1244467691 | 840 | V>A | No | TOPMed | |
| rs1229632135 | 841 | A>S | No | gnomAD | |
|
COSM1417071 rs200079670 |
842 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs772388012 | 843 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs2066190638 | 844 | N>D | No | TOPMed | |
| rs1300565978 | 846 | S>P | No | gnomAD | |
| rs746223663 | 849 | P>L | No |
ExAC gnomAD |
|
| rs2066189687 | 850 | E>Q | No | Ensembl | |
| rs1407238581 | 851 | R>C | No |
TOPMed gnomAD |
|
| rs199926321 | 851 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs199926321 | 851 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs199926321 | 851 | R>P | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1407238581 | 851 | R>S | No |
TOPMed gnomAD |
|
| COSM326193 | 852 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs755618323 | 854 | V>M | No |
ExAC gnomAD |
|
| rs1485437475 | 855 | S>A | No | gnomAD | |
| rs2066188410 | 855 | S>F | No | Ensembl | |
| rs759339200 | 856 | T>I | No |
ExAC TOPMed gnomAD |
|
| rs759339200 | 856 | T>S | No |
ExAC TOPMed gnomAD |
|
| rs766337040 | 857 | R>Q | No |
ExAC gnomAD |
|
| rs751289776 | 857 | R>W | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 858 | I>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1601699924 | 858 | I>V | No | Ensembl | |
| rs780634166 | 859 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs373347520 | 862 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs369024095 | 863 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs369024095 | 863 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
|
rs758252261 COSM1417070 |
865 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs2066167309 | 866 | E>* | No | Ensembl | |
| rs2066167165 | 866 | E>G | No | TOPMed | |
| rs2147413465 | 867 | T>A | No | Ensembl | |
| rs200847817 | 867 | T>M | No |
ESP ExAC gnomAD |
|
| rs141252891 | 868 | P>L | No | Ensembl | |
| rs1275405215 | 868 | P>S | No | TOPMed | |
| rs1432723103 | 869 | F>L | No |
TOPMed gnomAD |
|
| rs369732594 | 870 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1441303500 | 871 | G>E | No | gnomAD | |
| rs763230043 | 872 | G>D | No |
ExAC TOPMed gnomAD |
|
| rs1180607449 | 872 | G>R | No |
TOPMed gnomAD |
|
| rs763230043 | 872 | G>V | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 872 | G>V | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM5090428 | 873 | V>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2066164664 | 873 | V>I | No | TOPMed | |
| rs372327269 | 874 | E>K | No |
ESP ExAC TOPMed gnomAD |
|
| rs372327269 | 874 | E>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1217878121 | 875 | V>G | No | gnomAD | |
| rs368564765 | 877 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2066163751 | 878 | F>S | No | Ensembl | |
| rs779699565 | 879 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs1413009020 | 881 | L>P | No | gnomAD | |
| TCGA novel | 881 | L>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs771691320 | 881 | L>V | No |
ExAC gnomAD |
|
| rs200111358 | 883 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs201999293 | 883 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3842936 rs1382460521 |
884 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs1012249197 | 886 | P>L | No | gnomAD | |
| rs1012249197 | 886 | P>R | No | gnomAD | |
| rs200791148 | 887 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs2066161416 | 887 | N>Y | No | Ensembl | |
| rs1809975386 | 888 | V>A | No | TOPMed | |
| TCGA novel | 888 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1474585625 | 889 | Q>* | No | gnomAD | |
| rs766737874 | 889 | Q>H | No |
ExAC gnomAD |
|
| COSM1035410 | 890 | F>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769690739 | 896 | K>Q | No |
ExAC TOPMed gnomAD |
|
| rs1191997177 | 898 | L>F | No | gnomAD | |
| rs1191997177 | 898 | L>V | No | gnomAD | |
| rs1490829903 | 901 | E>G | No | Ensembl | |
| rs748136803 | 901 | E>K | No |
1000Genomes ExAC gnomAD |
|
| rs780989066 | 902 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs1436129405 | 905 | G>R | No |
TOPMed gnomAD |
|
| rs745882011 | 906 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs745882011 | 906 | P>Q | No |
ExAC TOPMed gnomAD |
|
| rs1207839574 | 906 | P>S | No | Ensembl | |
| rs995047869 | 908 | A>T | No | Ensembl | |
| rs754227017 | 908 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1439447530 | 910 | G>A | No |
TOPMed gnomAD |
|
| rs1439447530 | 910 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs753303246 | 910 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs753303246 | 910 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs2066140452 | 911 | T>P | No | TOPMed | |
| rs1334623640 | 912 | T>A | No | gnomAD | |
| rs1328796672 | 912 | T>I | No | gnomAD | |
| rs951133275 | 914 | T>A | No | Ensembl | |
| rs951133275 | 914 | T>P | No | Ensembl | |
| rs767850259 | 914 | T>S | No |
ExAC gnomAD |
|
| rs1166598462 | 915 | I>F | No | TOPMed | |
| COSM1035408 | 916 | H>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1377653283 | 917 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 918 | T>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2066138586 | 919 | H>Q | No | TOPMed | |
| rs868423887 | 922 | T>M | No |
TOPMed gnomAD |
|
| rs2066137692 | 924 | S>F | No | Ensembl | |
| rs1468688470 | 925 | Q>R | No |
TOPMed gnomAD |
|
| TCGA novel | 928 | V>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs769779572 | 928 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs923565565 | 929 | R>L | No |
TOPMed gnomAD |
|
| rs923565565 | 929 | R>Q | No |
TOPMed gnomAD |
|
| rs747944816 | 929 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs75735238 | 930 | V>L | No | Ensembl | |
| rs2066136321 | 931 | T>A | No | TOPMed | |
| rs201547301 | 933 | N>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs778814426 | 934 | G>D | No | ExAC | |
| rs141762500 | 934 | G>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs62621372 | 935 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs777954936 | 936 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs375115998 | 938 | K>E | No |
ESP gnomAD |
|
| rs755337831 | 940 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2066116871 | 942 | F>L | No | Ensembl | |
| rs2066116557 | 943 | G>R | No | Ensembl | |
| rs149805063 | 944 | A>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs149805063 | 944 | A>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1392377758 | 944 | A>T | No | gnomAD | |
|
rs149805063 RCV000888880 |
944 | A>V | No |
ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
| rs749888275 | 947 | Q>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 948 | C>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1166985607 | 950 | T>A | No | TOPMed | |
| rs2066115011 | 952 | P>S | No | TOPMed | |
| rs2066114883 | 953 | Q>* | No | TOPMed | |
| rs927746358 | 953 | Q>P | No | gnomAD | |
| rs927746358 | 953 | Q>R | No | gnomAD | |
| rs1364327455 | 954 | A>T | No |
TOPMed gnomAD |
|
| rs370055566 | 954 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs375999957 | 956 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs763839691 | 956 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs775416355 | 957 | G>D | No |
ExAC gnomAD |
|
| rs2066113215 | 957 | G>S | No | TOPMed | |
| rs1253631236 | 958 | Q>R | No | gnomAD | |
| rs200474256 | 959 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs62621407 | 959 | M>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2066112175 | 959 | M>R | No | Ensembl | |
| rs62621407 | 959 | M>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1281407079 | 960 | L>F | No | gnomAD | |
| rs1281407079 | 960 | L>V | No | gnomAD | |
| rs773225348 | 961 | L>V | No |
ExAC gnomAD |
|
| rs1392075655 | 963 | V>F | No | gnomAD | |
| rs1601697169 | 963 | V>G | No | Ensembl | |
| rs2147401738 | 964 | S>F | No | Ensembl | |
| TCGA novel | 965 | Y>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs558571072 | 966 | G>R | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs747342576 | 967 | G>A | No |
ExAC TOPMed gnomAD |
|
| rs747342576 | 967 | G>E | No |
ExAC TOPMed gnomAD |
|
|
rs745397200 COSM3555696 |
968 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 968 | S>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs569852454 | 970 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1007779940 | 971 | P>S | No | Ensembl | |
| rs2147401382 | 972 | N>T | No | Ensembl | |
| rs1420382741 | 973 | P>H | No |
TOPMed gnomAD |
|
| rs1420382741 | 973 | P>L | No |
TOPMed gnomAD |
|
| rs1483052137 | 974 | G>D | No | gnomAD | |
| rs201704027 | 974 | G>R | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201704027 | 974 | G>S | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1213523378 | 976 | F>I | No |
TOPMed gnomAD |
|
| rs767463618 | 977 | F>Y | No |
ExAC gnomAD |
|
| rs2147400975 | 978 | T>I | No | Ensembl | |
| rs774240975 | 978 | T>P | No |
ExAC gnomAD |
|
| rs765299932 | 979 | Y>S | No |
ExAC gnomAD |
|
| rs201751886 | 980 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs79500228 | 980 | R>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs79500228 | 980 | R>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201751886 | 980 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs775951287 | 981 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs202201273 | 982 | N>T | No |
ExAC gnomAD |
|
| rs748762533 | 984 | V>E | No |
ExAC gnomAD |
|
| rs200118373 | 984 | V>I | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200118373 | 984 | V>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1258871941 | 986 | R>* | No |
TOPMed gnomAD |
|
| rs1258871941 | 986 | R>G | No |
TOPMed gnomAD |
|
| rs201068347 | 986 | R>Q | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs957289874 | 987 | A>G | No |
TOPMed gnomAD |
|
| rs957289874 | 987 | A>V | No |
TOPMed gnomAD |
|
| rs1255836198 | 988 | F>C | No | gnomAD | |
| rs202117989 | 988 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
| rs754762851 | 989 | E>K | No |
ExAC gnomAD |
|
| rs754762851 | 989 | E>Q | No |
ExAC gnomAD |
|
| COSM5859677 | 990 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM78321 | 990 | P>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs766283296 | 992 | R>* | No |
ExAC gnomAD |
|
| rs375104267 | 992 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1339663316 | 995 | A>T | No | gnomAD | |
|
rs1189271682 COSM1221300 |
996 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic TOPMed |
| rs1189271682 | 996 | S>T | No | TOPMed | |
| rs1335784939 | 999 | R>C | No | gnomAD | |
| rs769351345 | 999 | R>H | No |
ExAC gnomAD |
|
| rs2066051900 | 1000 | S>G | No | gnomAD | |
| rs373694108 | 1003 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs746803740 | 1004 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1392481472 | 1006 | Q>E | No |
TOPMed gnomAD |
|
| rs2066050953 | 1009 | S>G | No | TOPMed | |
| rs2066050764 | 1009 | S>N | No | TOPMed | |
| rs750444951 | 1012 | Q>H | No |
ExAC TOPMed gnomAD |
|
| rs1468364555 | 1012 | Q>K | No |
TOPMed gnomAD |
|
| rs2066049878 | 1013 | R>G | No | Ensembl | |
| rs778706365 | 1013 | R>S | No |
ExAC TOPMed gnomAD |
|
| rs752781092 | 1016 | M>V | No |
ExAC TOPMed gnomAD |
|
| rs1569162645 | 1017 | V>A | No | Ensembl | |
| rs767871551 | 1017 | V>M | No |
ExAC gnomAD |
|
| rs766832996 | 1019 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs1213702854 | 1019 | I>V | No | Ensembl | |
| rs374289741 | 1020 | A>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs201274338 | 1020 | A>T | No |
TOPMed gnomAD |
|
| rs374289741 | 1020 | A>V | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs765665766 | 1021 | E>A | No |
ExAC gnomAD |
|
| rs1318244384 | 1021 | E>D | No |
TOPMed gnomAD |
|
| rs761344667 | 1022 | P>S | No |
ExAC TOPMed gnomAD |
|
| rs1377598598 | 1024 | Q>P | No | gnomAD | |
| rs768134621 | 1025 | S>F | No |
ExAC gnomAD |
|
| rs1414053033 | 1026 | W>S | No | gnomAD | |
| TCGA novel | 1027 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs576902370 | 1028 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs771965276 | 1029 | P>L | No |
ExAC gnomAD |
|
| rs368989847 | 1030 | R>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs763070819 | 1030 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs368989847 | 1030 | R>W | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs970801068 | 1032 | A>S | No | Ensembl | |
| rs2066045819 | 1033 | E>K | No | TOPMed | |
| rs543422894 | 1034 | S>C | No | 1000Genomes | |
| rs2147389860 | 1034 | S>P | No | Ensembl | |
| rs755001480 | 1035 | L>V | No |
1000Genomes ExAC gnomAD |
|
| rs1285892314 | 1036 | Q>* | No | gnomAD | |
| rs11547728 | 1036 | Q>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
|
COSM3842935 rs1345121299 |
1037 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| rs766445738 | 1038 | M>T | No |
ExAC TOPMed gnomAD |
|
|
COSM5366523 rs372167877 |
1039 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs865841168 | 1040 | V>M | No | Ensembl | |
| rs914823000 | 1041 | V>A | No |
TOPMed gnomAD |
|
| rs914823000 | 1041 | V>G | No |
TOPMed gnomAD |
|
| rs1028659143 | 1042 | G>R | No |
TOPMed gnomAD |
|
| rs373669856 | 1045 | Y>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs1292876817 | 1046 | V>L | No |
TOPMed gnomAD |
|
| rs1292876817 | 1046 | V>M | No |
TOPMed gnomAD |
|
| rs1356200724 | 1048 | H>L | No |
TOPMed gnomAD |
|
| rs1166542517 | 1049 | N>D | No |
TOPMed gnomAD |
|
| rs1428841737 | 1049 | N>S | No |
TOPMed gnomAD |
|
| rs1292222969 | 1051 | T>S | No | gnomAD | |
| rs534785502 | 1054 | V>F | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs534785502 | 1054 | V>I | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2066030629 | 1055 | F>L | No | TOPMed | |
| TCGA novel | 1056 | L>C | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM3723108 | 1057 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs773101215 | 1058 | P>L | No |
ExAC TOPMed gnomAD |
|
| COSM3390305 | 1058 | P>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2066029700 | 1059 | A>V | No | Ensembl | |
| rs761703447 | 1061 | P>L | No |
ExAC gnomAD |
|
| rs769599289 | 1061 | P>S | No |
ExAC gnomAD |
|
| rs1300753731 | 1066 | A>T | No | gnomAD | |
| rs373860676 | 1067 | Y>H | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779118429 | 1069 | L>F | No |
ExAC TOPMed gnomAD |
|
| rs1375773688 | 1070 | T>K | No |
TOPMed gnomAD |
|
| rs1375773688 | 1070 | T>M | No |
TOPMed gnomAD |
|
| rs1213754705 | 1071 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs371203017 | 1073 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs139926938 | 1074 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2147386428 | 1075 | M>T | No | Ensembl | |
| rs2066026614 | 1077 | G>R | No |
TOPMed gnomAD |
|
| rs374627819 | 1079 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs374627819 | 1079 | R>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs200429266 | 1079 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs200429266 | 1079 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs374627819 | 1079 | R>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1244302654 | 1080 | A>P | No |
TOPMed gnomAD |
|
| rs1244302654 | 1080 | A>S | No |
TOPMed gnomAD |
|
| rs1160131860 | 1080 | A>V | No | gnomAD | |
| rs1258571413 | 1084 | T>P | No | gnomAD | |
| rs1202536398 | 1085 | E>G | No | gnomAD | |
| rs766161309 | 1086 | A>T | No |
ExAC gnomAD |
|
| rs758165010 | 1086 | A>V | No |
ExAC gnomAD |
|
| rs534307672 | 1087 | G>R | No |
ExAC TOPMed gnomAD |
|
| rs946043122 | 1088 | A>D | No | Ensembl | |
| rs373329714 | 1088 | A>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs946043122 | 1088 | A>V | No | Ensembl | |
| rs759629858 | 1090 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs759629858 | 1090 | E>Q | No |
ExAC TOPMed gnomAD |
|
| rs770979139 | 1092 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs770979139 | 1092 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1569162048 | 1093 | P>R | No | Ensembl | |
| rs749573114 | 1093 | P>S | No |
ExAC gnomAD |
|
| rs372865746 | 1094 | D>E | No |
ESP ExAC gnomAD |
|
| rs2066022552 | 1094 | D>N | No | TOPMed | |
| rs2066021969 | 1095 | P>S | No | gnomAD | |
| rs1454918774 | 1096 | T>I | No |
TOPMed gnomAD |
|
| rs1454918774 | 1096 | T>N | No |
TOPMed gnomAD |
|
| rs530361396 | 1100 | F>L | No |
1000Genomes ExAC gnomAD |
|
| rs974464419 | 1102 | G>D | No | TOPMed | |
| rs747486856 | 1104 | V>F | No |
ExAC TOPMed gnomAD |
|
| rs747486856 | 1104 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs747486856 | 1104 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs2066020341 | 1106 | K>E | No | Ensembl | |
| rs779759334 | 1107 | Q>R | No |
ExAC gnomAD |
|
| rs1205890572 | 1114 | A>T | No |
TOPMed gnomAD |
|
| rs764874526 | 1115 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1266389693 | 1115 | R>W | No |
TOPMed gnomAD |
|
| rs776979346 | 1118 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs769207954 | 1121 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 1122 | A>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs375577050 | 1122 | A>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs375577050 | 1122 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1209043498 | 1123 | M>T | No | gnomAD | |
| rs772579863 | 1124 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2066004358 | 1125 | L>P | No | TOPMed | |
| rs778521030 | 1126 | Q>P | No |
ExAC TOPMed gnomAD |
|
| rs749040680 | 1129 | E>K | No |
ExAC TOPMed gnomAD |
|
| COSM1417067 | 1132 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs751429886 | 1135 | E>K | No |
ExAC gnomAD |
|
| TCGA novel | 1135 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1307657362 | 1136 | R>C | No | gnomAD | |
| rs377737626 | 1136 | R>H | No |
ESP ExAC TOPMed gnomAD |
|
| rs377737626 | 1136 | R>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1170945805 | 1138 | T>I | No |
TOPMed gnomAD |
|
| rs1312222476 | 1138 | T>S | No | gnomAD | |
| rs1170945805 | 1138 | T>S | No |
TOPMed gnomAD |
|
| rs374311597 | 1139 | M>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1184195892 | 1139 | M>T | No | gnomAD | |
| rs1364345446 | 1139 | M>V | No | gnomAD | |
| rs2066001865 | 1140 | K>* | No | Ensembl | |
| TCGA novel | 1141 | T>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1250714562 | 1141 | T>M | No |
TOPMed gnomAD |
|
|
COSM1035406 rs1443835685 |
1143 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1244964018 | 1145 | T>I | No |
TOPMed gnomAD |
|
| rs772289683 | 1146 | D>H | No |
ExAC gnomAD |
|
| rs1230700141 | 1149 | C>Y | No | gnomAD | |
| rs1288185100 | 1152 | P>L | No | gnomAD | |
| rs2065999564 | 1153 | E>G | No | TOPMed | |
| rs777426414 | 1154 | V>L | No |
ExAC gnomAD |
|
| rs777426414 | 1154 | V>M | No |
ExAC gnomAD |
|
| COSM3424269 | 1156 | P>A | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs953456826 | 1156 | P>H | No | Ensembl | |
| rs376149647 | 1157 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2065999099 | 1157 | P>S | No | Ensembl | |
| rs1389397362 | 1159 | K>N | No |
TOPMed gnomAD |
|
| rs1471607896 | 1160 | R>Q | No |
TOPMed gnomAD |
|
| rs781014569 | 1160 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1182273624 | 1161 | R>Q | No | gnomAD | |
| rs754714515 | 1161 | R>W | No |
ExAC TOPMed gnomAD |
|
| COSM3800286 | 1162 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2065997243 | 1163 | K>Q | No | TOPMed | |
| rs747034054 | 1164 | R>* | No |
ExAC gnomAD |
|
| rs373453904 | 1164 | R>Q | No |
ESP ExAC TOPMed gnomAD |
|
| rs1269542800 | 1165 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1269542800 | 1165 | D>Y | No | gnomAD | |
| rs2065996335 | 1167 | T>A | No | TOPMed | |
| rs753916806 | 1167 | T>R | No |
ExAC gnomAD |
|
| rs1381737303 | 1168 | H>R | No | gnomAD | |
| rs1229284132 | 1168 | H>Y | No | gnomAD | |
| rs1282658376 | 1171 | P>L | No |
TOPMed gnomAD |
|
| rs767996562 | 1172 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs752942511 | 1172 | E>K | No |
ExAC gnomAD |
|
| rs2065994792 | 1173 | F>L | No | TOPMed | |
| rs2065994435 | 1174 | I>M | No | gnomAD | |
| rs756696997 | 1174 | I>T | No |
ExAC TOPMed gnomAD |
|
| rs1431685062 | 1175 | V>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs201645775 | 1177 | F>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs745854880 | 1178 | G>S | No |
ExAC gnomAD |
|
| rs1192425761 | 1179 | S>A | No |
TOPMed gnomAD |
|
| rs1601691401 | 1179 | S>F | No | Ensembl | |
| rs372197536 | 1180 | R>C | No |
ESP TOPMed gnomAD |
|
| rs1455752970 | 1180 | R>H | No |
TOPMed gnomAD |
|
| rs770010469 | 1181 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2065985604 | 1183 | V>M | No | TOPMed | |
| rs375098920 | 1186 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs755195831 | 1186 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs1302135367 | 1187 | V>M | No |
TOPMed gnomAD |
|
| rs2065984513 | 1188 | E>Q | No | Ensembl | |
| rs750933296 | 1190 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs2065984062 | 1191 | T>A | No | TOPMed | |
| rs1345961455 | 1191 | T>I | No | Ensembl | |
| rs765673534 | 1192 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs1372118434 | 1192 | R>W | No |
TOPMed gnomAD |
|
| rs536523204 | 1193 | V>M | No |
1000Genomes ExAC gnomAD |
|
| rs763625119 | 1195 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs751845658 | 1196 | V>E | No | Ensembl | |
|
rs1423712452 COSM1035404 |
1196 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs371055458 | 1197 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs774456849 | 1200 | L>F | No |
ExAC gnomAD |
|
| rs190470503 | 1203 | P>L | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1425765342 | 1204 | L>P | No | TOPMed | |
| rs2065981323 | 1206 | I>V | No | Ensembl | |
| rs747263348 | 1207 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs747263348 | 1207 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs2065980800 | 1208 | P>R | No | TOPMed | |
| rs1435253814 | 1211 | V>I | No | Ensembl | |
| rs763081231 | 1212 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs763081231 | 1212 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs150309152 | 1213 | I>V | No |
1000Genomes TOPMed gnomAD |
|
| rs370284603 | 1214 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs754343905 | 1214 | A>V | No |
ExAC TOPMed gnomAD |
|
|
COSM4831460 rs760225048 |
1215 | V>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC TOPMed gnomAD NCI-TCGA Cosmic |
| rs760225048 | 1215 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1320166252 | 1216 | S>A | No | TOPMed | |
| rs1221057887 | 1216 | S>C | No |
TOPMed gnomAD |
|
| rs1358003534 | 1217 | V>A | No | gnomAD | |
| rs1458279706 | 1218 | Y>N | No | gnomAD | |
| rs371130944 | 1218 | Y>S | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs759439955 | 1219 | C>W | No |
ExAC TOPMed gnomAD |
|
| rs1014538178 | 1220 | Y>F | No | gnomAD | |
| rs1014538178 | 1220 | Y>S | No | gnomAD | |
| rs2065977604 | 1221 | W>R | No | Ensembl | |
| rs778176942 | 1223 | K>R | No |
ExAC gnomAD |
|
| rs369828232 | 1227 | A>T | No |
ESP ExAC gnomAD |
|
| rs2065958882 | 1227 | A>V | No | TOPMed | |
| rs1224789123 | 1231 | Y>* | No | TOPMed | |
| rs1192542607 | 1232 | E>K | No | gnomAD | |
| rs888633390 | 1235 | K>N | No |
TOPMed gnomAD |
|
| rs1207870229 | 1236 | S>F | No | gnomAD | |
| COSM4406268 | 1237 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| COSM726732 | 1239 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2065956712 | 1239 | E>Q | No | Ensembl | |
| rs766000331 | 1240 | G>A | No |
ExAC gnomAD |
|
| rs751262783 | 1240 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1452915116 | 1248 | R>C | No |
TOPMed gnomAD |
|
| rs1332220132 | 1248 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs750191330 | 1249 | C>F | No |
ExAC gnomAD |
|
| TCGA novel | 1249 | C>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1249 | C>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM1035403 | 1250 | K>E | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1250 | K>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs764910604 | 1251 | K>R | No |
ExAC gnomAD |
|
| rs2065955362 | 1252 | E>* | No | TOPMed | |
| rs753759595 | 1255 | D>Y | No |
ExAC gnomAD |
|
| rs1464914163 | 1256 | L>V | No |
TOPMed gnomAD |
|
| rs750672460 | 1258 | I>M | No |
ExAC TOPMed gnomAD |
|
| rs2065941858 | 1258 | I>V | No | TOPMed | |
| rs765509654 | 1259 | E>K | No |
ExAC gnomAD |
|
| rs2065941396 | 1260 | M>I | No | Ensembl | |
| TCGA novel | 1261 | E>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs984384699 | 1263 | Q>H | No | Ensembl | |
| rs777175182 | 1263 | Q>K | No |
ExAC gnomAD |
|
| TCGA novel | 1265 | N>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1245369694 COSM1035402 |
1266 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs375409236 | 1267 | V>L | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM4855087 rs375409236 |
1267 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs952926750 | 1268 | H>Y | No | Ensembl | |
| COSM461056 | 1269 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| TCGA novel | 1269 | E>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065939253 | 1270 | A>T | No | TOPMed | |
| TCGA novel | 1270 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1383023136 | 1271 | G>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1200162281 | 1272 | I>V | No | gnomAD | |
| rs1601688609 | 1274 | V>A | No | Ensembl | |
| rs1251509632 | 1274 | V>M | No |
TOPMed gnomAD |
|
| rs748920566 | 1276 | D>G | No |
ExAC TOPMed gnomAD |
|
| rs771855846 | 1279 | T>I | No | gnomAD | |
| rs1242017173 | 1281 | T>I | No | gnomAD | |
| rs1368129076 | 1281 | T>S | No | TOPMed | |
| rs1453635304 | 1282 | D>N | No |
TOPMed gnomAD |
|
| rs201960199 | 1283 | R>C | No |
ESP ExAC TOPMed gnomAD |
|
| rs748120996 | 1283 | R>H | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs1212849563 |
1284 | V>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1212849563 | 1284 | V>L | No |
TOPMed gnomAD |
|
| TCGA novel | 1286 | F>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs374108984 | 1288 | P>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs1361223860 | 1288 | P>S | No |
TOPMed gnomAD |
|
| TCGA novel | 1289 | S>I | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs757601022 | 1292 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1000095053 | 1293 | D>N | No | gnomAD | |
| rs2065934687 | 1294 | K>E | No | Ensembl | |
| rs2147368973 | 1296 | V>A | No | Ensembl | |
| rs760877226 | 1296 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs776075016 | 1297 | M>I | No |
ExAC gnomAD |
|
| rs1601688478 | 1299 | T>P | No | Ensembl | |
| rs2065933545 | 1300 | G>A | No | TOPMed | |
| rs759115091 | 1300 | G>C | No |
ExAC TOPMed gnomAD |
|
| rs759115091 | 1300 | G>S | No |
ExAC TOPMed gnomAD |
|
| rs1267280267 | 1301 | K>N | No | gnomAD | |
| rs904437628 | 1304 | I>L | No | Ensembl | |
| rs1601688454 | 1304 | I>T | No | Ensembl | |
| rs559938153 | 1305 | P>S | No |
1000Genomes ExAC gnomAD |
|
| rs1601688422 | 1306 | E>G | No | Ensembl | |
| rs772789014 | 1306 | E>K | No |
ExAC gnomAD |
|
| rs772789014 | 1306 | E>Q | No |
ExAC gnomAD |
|
| rs201378041 | 1307 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1279245268 | 1307 | P>S | No |
TOPMed gnomAD |
|
| rs746941566 | 1308 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs754910456 | 1308 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs1057253566 | 1309 | R>L | No | gnomAD | |
| rs1057253566 | 1309 | R>Q | No | gnomAD | |
| rs1351986520 | 1309 | R>W | No |
TOPMed gnomAD |
|
| rs201776829 | 1310 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs200080652 | 1310 | P>T | No |
ESP ExAC TOPMed gnomAD |
|
| rs2065930337 | 1312 | V>L | No | Ensembl | |
| TCGA novel | 1316 | L>P | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748354677 | 1320 | S>A | No | Ensembl | |
| COSM1417065 | 1320 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1601688328 | 1321 | N>T | No | Ensembl | |
| TCGA novel | 1322 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs752961600 | 1325 | S>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1327 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1601688307 | 1328 | F>S | No | Ensembl | |
| rs1175630159 | 1330 | I>V | No | gnomAD | |
| rs2065928453 | 1331 | N>D | No | Ensembl | |
| rs200739841 | 1331 | N>I | No |
ExAC TOPMed gnomAD |
|
| rs200739841 | 1331 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs764718458 | 1334 | H>Q | No |
ExAC gnomAD |
|
| rs1287532583 | 1335 | T>N | No | gnomAD | |
| rs2065917853 | 1339 | Q>H | No | gnomAD | |
| rs1056972715 | 1340 | R>Q | No | gnomAD | |
| rs1453452385 | 1340 | R>W | No | gnomAD | |
| rs753441958 | 1341 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs2065917287 | 1341 | E>G | No | TOPMed | |
| TCGA novel | 1342 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs539295991 | 1343 | S>L | No |
1000Genomes TOPMed gnomAD |
|
| rs200418278 | 1344 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
|
rs775464863 COSM5859674 |
1345 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| COSM1417064 | 1345 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs769778490 | 1346 | A>P | No |
ExAC TOPMed gnomAD |
|
| rs769778490 | 1346 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs769778490 | 1346 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1259523188 | 1350 | F>L | No |
TOPMed gnomAD |
|
|
COSM3065306 rs1204353997 |
1351 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs776753345 | 1351 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs549095175 | 1355 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs753348363 | 1357 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1444434763 | 1360 | G>R | No | Ensembl | |
| TCGA novel | 1362 | L>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065914054 | 1363 | E>V | No | Ensembl | |
| rs767425660 | 1366 | T>A | No |
ExAC gnomAD |
|
| rs759348018 | 1366 | T>M | No |
ExAC TOPMed gnomAD |
|
| COSM380546 | 1367 | D>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs28499460 | 1368 | I>T | No | Ensembl | |
| rs762771999 | 1369 | M>I | No |
ExAC gnomAD |
|
| rs2065913105 | 1370 | H>R | No | Ensembl | |
| rs776823804 | 1371 | T>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1371 | T>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs775620720 | 1373 | F>L | No |
ExAC gnomAD |
|
| TCGA novel | 1373 | F>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs772435940 | 1375 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1336814368 | 1378 | E>K | No | gnomAD | |
| rs2065911591 | 1380 | Y>* | No | TOPMed | |
| rs755621229 | 1381 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs755621229 | 1381 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1285642310 | 1382 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1403460980 | 1383 | A>T | No | gnomAD | |
| rs2065910871 | 1384 | K>E | No | Ensembl | |
| rs73891209 | 1384 | K>N | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1411731682 | 1389 | M>L | No | gnomAD | |
|
rs1466531970 COSM3694137 |
1391 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
| rs1431309803 | 1391 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| COSM4105097 | 1392 | R>G | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs776342572 | 1393 | S>P | No |
ExAC gnomAD |
|
| COSM5499451 | 1396 | V>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768048613 | 1396 | V>L | No |
ExAC gnomAD |
|
| COSM419495 | 1399 | R>S | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1271811572 | 1400 | M>L | No |
TOPMed gnomAD |
|
| rs906534424 | 1402 | S>C | No | TOPMed | |
| rs906534424 | 1402 | S>Y | No | TOPMed | |
| rs778648306 | 1403 | N>S | No |
ExAC TOPMed gnomAD |
|
| rs757234543 | 1405 | M>T | No |
ExAC gnomAD |
|
| COSM1308290 | 1407 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1292126822 | 1409 | L>V | No |
TOPMed gnomAD |
|
| rs2065877859 | 1411 | Q>P | No | TOPMed | |
| rs1474711307 | 1413 | L>V | No | gnomAD | |
| rs1202488608 | 1415 | D>N | No | gnomAD | |
| rs1263627580 | 1416 | S>G | No | gnomAD | |
| rs2147351814 | 1417 | A>T | No | Ensembl | |
|
TCGA novel rs2147351798 |
1417 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs1321070645 | 1418 | G>R | No | gnomAD | |
| COSM6162464 | 1419 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2065858233 | 1419 | E>D | No | Ensembl | |
| rs1056628857 | 1419 | E>K | No |
TOPMed gnomAD |
|
| rs1056628857 | 1419 | E>Q | No |
TOPMed gnomAD |
|
| rs2147351653 | 1420 | P>S | No | Ensembl | |
| rs2065857818 | 1423 | K>R | No | Ensembl | |
| rs374294294 | 1424 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM3842932 rs1300644254 |
1425 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA Cosmic |
| rs1440787898 | 1428 | I>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs2147351482 | 1428 | I>T | No | Ensembl | |
| rs766461191 | 1429 | K>R | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1430 | H>A | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs758755240 | 1430 | H>N | No |
ExAC gnomAD |
|
| rs758755240 | 1430 | H>Y | No |
ExAC gnomAD |
|
| rs2147351385 | 1431 | Q>* | No | Ensembl | |
| rs1471238582 | 1431 | Q>H | No | gnomAD | |
| rs2147351297 | 1434 | K>M | No | Ensembl | |
| rs2147351297 | 1434 | K>R | No | Ensembl | |
| rs2147351297 | 1434 | K>T | No | Ensembl | |
| rs1332845035 | 1436 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs765703084 | 1437 | V>M | No |
ExAC gnomAD |
|
| rs906945700 | 1438 | D>G | No | Ensembl | |
| rs2147351169 | 1438 | D>N | No | Ensembl | |
| rs1410388747 | 1439 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs2147351099 | 1440 | V>I | No | Ensembl | |
| rs1232704305 | 1441 | Q>* | No |
TOPMed gnomAD |
|
| rs2065855353 | 1442 | K>E | No | TOPMed | |
| rs2065855226 | 1442 | K>R | No | TOPMed | |
| rs370462169 | 1443 | K>* | No |
ESP ExAC TOPMed gnomAD |
|
| rs370462169 | 1443 | K>E | No |
ESP ExAC TOPMed gnomAD |
|
| rs764463882 | 1443 | K>M | No |
ExAC TOPMed gnomAD |
|
| rs764463882 | 1443 | K>T | No |
ExAC TOPMed gnomAD |
|
| rs761210103 | 1444 | A>V | No |
ExAC TOPMed gnomAD |
|
| rs1474596231 | 1446 | Y>C | No | Ensembl | |
| rs771557333 | 1447 | T>A | No |
ExAC gnomAD |
|
|
rs376965540 COSM3555685 |
1448 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs376965540 | 1448 | L>V | No |
ESP ExAC TOPMed gnomAD |
|
| rs770735940 | 1450 | D>N | No |
ExAC TOPMed gnomAD |
|
| rs748998767 | 1451 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2147350630 | 1452 | G>W | No | Ensembl | |
| rs2147350535 | 1454 | L>R | No | Ensembl | |
| rs2147350468 | 1455 | G>E | No | Ensembl | |
| rs1302662221 | 1455 | G>R | No | gnomAD | |
| rs2147350449 | 1456 | D>N | No | Ensembl | |
| rs2147350449 | 1456 | D>Y | No | Ensembl | |
| rs372967126 | 1458 | V>A | No |
ESP ExAC TOPMed gnomAD |
|
| rs780069040 | 1460 | Y>* | No |
ExAC TOPMed gnomAD |
|
| rs758488420 | 1461 | A>T | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1463 | L>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065851633 | 1463 | L>V | No | Ensembl | |
| rs764957819 | 1464 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs2065803638 | 1464 | T>S | No | gnomAD | |
| rs1385164203 | 1465 | V>A | No | gnomAD | |
| rs1305130308 | 1465 | V>L | No | gnomAD | |
| rs768751941 | 1466 | S>C | No |
ExAC TOPMed gnomAD |
|
| rs1050704647 | 1466 | S>R | No |
TOPMed gnomAD |
|
| rs1233904904 | 1466 | S>T | No |
TOPMed gnomAD |
|
| rs1601680133 | 1467 | V>G | No | Ensembl | |
| rs746916579 | 1467 | V>L | No |
ExAC TOPMed gnomAD |
|
| rs746916579 | 1467 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs998098306 | 1468 | I>S | No | Ensembl | |
| rs998098306 | 1468 | I>T | No | Ensembl | |
| rs774626695 | 1468 | I>V | No |
ExAC TOPMed gnomAD |
|
| rs2065801214 | 1469 | V>E | No |
TOPMed gnomAD |
|
| rs771037434 | 1469 | V>M | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1470 | Q>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065800920 | 1470 | Q>H | No | TOPMed | |
| rs1249297625 | 1470 | Q>R | No |
TOPMed gnomAD |
|
| rs777991567 | 1472 | E>K | No |
ExAC gnomAD |
|
| rs2065800372 | 1473 | G>E | No | TOPMed | |
| rs748554523 | 1476 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs1415622042 | 1478 | P>A | No |
TOPMed gnomAD |
|
|
rs200623878 COSM479115 |
1478 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs375377088 | 1479 | V>L | No |
ESP ExAC gnomAD |
|
| rs371453551 | 1480 | K>R | No |
ESP TOPMed gnomAD |
|
| COSM726734 | 1483 | N>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2065798686 | 1486 | T>A | No | TOPMed | |
| rs750949109 | 1488 | S>F | No |
TOPMed gnomAD |
|
| rs375291732 | 1490 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs2147341289 | 1493 | K>R | No | Ensembl | |
| COSM1417061 | 1493 | K>missing | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs764907177 | 1495 | I>S | No |
ExAC gnomAD |
|
| rs764907177 | 1495 | I>T | No |
ExAC gnomAD |
|
| rs1295164975 | 1495 | I>V | No |
TOPMed gnomAD |
|
| COSM1417060 | 1496 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1228305344 | 1498 | V>L | No |
TOPMed gnomAD |
|
| rs1569158038 | 1500 | R>G | No | gnomAD | |
| rs1470249034 | 1500 | R>H | No | gnomAD | |
| rs2065796632 | 1501 | G>R | No | TOPMed | |
| rs1183386920 | 1505 | S>F | No | gnomAD | |
| rs760760853 | 1505 | S>P | No |
ExAC gnomAD |
|
| rs1236891672 | 1507 | W>* | No | gnomAD | |
| rs1435511754 | 1507 | W>R | No | gnomAD | |
| COSM4105095 | 1509 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs2065795403 | 1511 | D>E | No | Ensembl | |
| rs770930229 | 1512 | S>R | No |
ExAC gnomAD |
|
| rs371319008 | 1513 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs2147338161 | 1517 | W>C | No | Ensembl | |
|
rs2065781631 TCGA novel |
1518 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1320459935 | 1518 | R>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed |
| rs1227731618 | 1519 | P>L | No | gnomAD | |
| rs2147338021 | 1520 | G>C | No | Ensembl | |
| TCGA novel | 1520 | G>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1271760642 | 1521 | S>F | No | gnomAD | |
| rs1397852647 | 1522 | T>A | No | gnomAD | |
| TCGA novel | 1522 | T>G | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065780810 | 1525 | I>T | No | Ensembl | |
| rs199650688 | 1527 | S>L | No |
ESP ExAC TOPMed gnomAD |
|
| rs2065779897 | 1532 | T>M | No | TOPMed | |
| rs1173302787 | 1534 | Q>R | No | gnomAD | |
| rs765375689 | 1535 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs751388537 | 1535 | R>W | No |
ExAC TOPMed gnomAD |
|
| rs761835002 | 1536 | E>V | No |
ExAC TOPMed gnomAD |
|
| rs1253547739 | 1537 | G>D | No |
TOPMed gnomAD |
|
| rs760860292 | 1538 | R>P | No |
ExAC gnomAD |
|
| rs760860292 | 1538 | R>Q | No |
ExAC gnomAD |
|
| rs182970183 | 1538 | R>W | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1274952446 | 1539 | W>* | No | gnomAD | |
| rs2065778164 | 1540 | K>E | No | TOPMed | |
| rs772316395 | 1540 | K>N | No |
ExAC gnomAD |
|
| rs746387640 | 1541 | R>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs779228197 | 1541 | R>H | No |
ExAC TOPMed gnomAD |
|
| rs201459786 | 1542 | V>I | No |
ESP ExAC TOPMed gnomAD |
|
| rs1355639445 | 1546 | M>L | No | gnomAD | |
| COSM3555683 | 1546 | M>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1020608286 | 1550 | V>A | No | Ensembl | |
| rs746824032 | 1551 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
COSM4388812 rs1252245593 |
1551 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed gnomAD |
| rs2065768959 | 1553 | G>R | No | Ensembl | |
| rs1482115584 | 1553 | G>V | No | gnomAD | |
| rs1333570512 | 1556 | L>F | No | TOPMed | |
| rs750351888 | 1560 | K>* | No |
ExAC gnomAD |
|
| rs764161574 | 1560 | K>N | No |
ExAC gnomAD |
|
| rs2065767807 | 1561 | V>L | No | Ensembl | |
| rs2065767807 | 1561 | V>M | No | Ensembl | |
| rs1376846095 | 1562 | G>R | No |
TOPMed gnomAD |
|
| rs2065767058 | 1564 | S>C | No |
TOPMed gnomAD |
|
| rs78482989 | 1565 | Q>E | No |
TOPMed gnomAD |
|
| rs78482989 | 1565 | Q>K | No |
TOPMed gnomAD |
|
| rs971202428 | 1566 | Q>L | No |
TOPMed gnomAD |
|
| rs368368664 | 1567 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| rs1258381746 | 1568 | E>Q | No | TOPMed | |
| rs1601678196 | 1569 | D>A | No | Ensembl | |
| rs1156953455 | 1570 | S>R | No |
TOPMed gnomAD |
|
| rs2147334828 | 1572 | Q>H | No | Ensembl | |
| rs2065765441 | 1573 | D>E | No | Ensembl | |
| rs991130214 | 1575 | P>L | No |
TOPMed gnomAD |
|
| rs2147334729 | 1575 | P>T | No | Ensembl | |
| rs2065764729 | 1576 | G>A | No |
TOPMed gnomAD |
|
| rs759773127 | 1578 | R>C | No |
ExAC TOPMed gnomAD |
|
| rs1389925655 | 1578 | R>L | No |
TOPMed gnomAD |
|
| rs1389925655 | 1578 | R>P | No |
TOPMed gnomAD |
|
| rs759231441 | 1579 | H>Y | No | Ensembl | |
| rs765707742 | 1582 | L>M | No |
ExAC gnomAD |
|
| rs2065751238 | 1583 | E>K | No | Ensembl | |
| rs1207476773 | 1584 | E>K | No |
TOPMed gnomAD |
|
| rs986307397 | 1584 | E>V | No | TOPMed | |
| rs1467120511 | 1585 | E>G | No | gnomAD | |
| rs1464739619 | 1586 | N>K | No |
TOPMed gnomAD |
|
| TCGA novel | 1586 | N>T | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs762362721 | 1587 | R>G | No |
ExAC gnomAD |
|
| rs776067354 | 1587 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs776067354 | 1587 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs762362721 | 1587 | R>W | No |
ExAC gnomAD |
|
| TCGA novel | 1590 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM4105093 | 1593 | R>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs768203410 | 1593 | R>P | No |
ExAC gnomAD |
|
| rs1431136184 | 1594 | P>A | No |
TOPMed gnomAD |
|
| rs760320759 | 1594 | P>L | No |
ExAC TOPMed gnomAD |
|
| rs2065748835 | 1595 | T>I | No | TOPMed | |
| rs879151685 | 1595 | T>P | No | Ensembl | |
| rs1199898952 | 1596 | D>N | No |
TOPMed gnomAD |
|
| rs2065748322 | 1596 | D>V | No |
TOPMed gnomAD |
|
| rs1601677213 | 1597 | E>* | No | Ensembl | |
|
TCGA novel rs1601677213 |
1597 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
| rs2065747702 | 1598 | V>L | No | Ensembl | |
| rs1276759347 | 1599 | D>G | No | gnomAD | |
| rs1346012264 | 1599 | D>N | No | gnomAD | |
| rs2065746948 | 1600 | E>Q | No | gnomAD | |
| rs991246172 | 1601 | G>C | No | TOPMed | |
| rs749235325 | 1604 | K>N | No |
ExAC gnomAD |
|
| rs781136241 | 1605 | R>K | No |
ExAC gnomAD |
|
| rs966470315 | 1606 | G>V | No | Ensembl | |
| rs201644992 | 1607 | S>R | No |
ESP ExAC TOPMed gnomAD |
|
| rs574952389 | 1608 | V>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1300839153 | 1610 | E>G | No | TOPMed | |
| rs1403924468 | 1611 | K>N | No | gnomAD | |
| rs1170507545 | 1612 | E>D | No |
TOPMed gnomAD |
|
| rs770516365 | 1613 | R>Q | No |
ExAC TOPMed gnomAD |
|
|
TCGA novel rs1465229293 |
1613 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs541570546 | 1614 | T>M | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs2065743539 | 1615 | K>E | No | Ensembl | |
| rs1232695446 | 1615 | K>T | No | gnomAD | |
| rs762298147 | 1616 | A>S | No |
ExAC gnomAD |
|
| rs1198992174 | 1617 | I>T | No | gnomAD | |
| rs2065742264 | 1621 | Y>C | No | gnomAD | |
| rs1351428481 | 1623 | T>M | No | gnomAD | |
| rs1448652231 | 1624 | R>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| rs1448652231 | 1624 | R>Q | No |
TOPMed gnomAD |
|
|
rs1275496634 COSM3065270 |
1624 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA TOPMed |
| TCGA novel | 1627 | S>F | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1627 | S>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1394044690 | 1629 | K>R | No | gnomAD | |
| rs1296480979 | 1636 | V>M | No |
TOPMed gnomAD |
|
| rs1401533379 | 1637 | D>H | No | gnomAD | |
| rs2065725558 | 1638 | N>S | No | Ensembl | |
| rs1348119470 | 1639 | F>C | No | TOPMed | |
| rs1173243448 | 1639 | F>L | No | gnomAD | |
| rs866299424 | 1640 | F>C | No | Ensembl | |
| rs866299424 | 1640 | F>Y | No | Ensembl | |
| TCGA novel | 1641 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1476977067 | 1642 | S>N | No | gnomAD | |
| TCGA novel | 1644 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1644 | L>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs778176391 | 1645 | A>V | No |
ExAC gnomAD |
|
| rs762034126 | 1646 | P>L | No | gnomAD | |
| rs762034126 | 1646 | P>R | No | gnomAD | |
| rs748677291 | 1647 | G>R | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| rs2065723269 | 1648 | H>P | No | gnomAD | |
| rs781634312 | 1649 | A>T | No |
ExAC TOPMed gnomAD |
|
| rs367781721 | 1649 | A>V | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs779415007 | 1650 | V>M | No |
ExAC TOPMed gnomAD |
|
| rs1454891870 | 1651 | P>R | No |
TOPMed gnomAD |
|
| rs187332808 | 1652 | P>L | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs758133473 | 1652 | P>S | No |
ExAC gnomAD |
|
| COSM3842931 | 1653 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs765133194 | 1653 | A>V | No |
ExAC gnomAD |
|
| rs2147326505 | 1654 | V>F | No | Ensembl | |
| COSM6095530 | 1657 | F>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs372267325 | 1658 | F>C | No |
ESP ExAC gnomAD |
|
| rs774549808 | 1658 | F>L | No |
ExAC TOPMed gnomAD |
|
| rs1320343171 | 1659 | D>N | No |
TOPMed gnomAD |
|
| rs1320343171 | 1659 | D>Y | No |
TOPMed gnomAD |
|
| rs1372120948 | 1660 | F>Y | No | TOPMed | |
| rs1438187906 | 1661 | L>P | No | TOPMed | |
| COSM6162465 | 1662 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs763241920 | 1663 | E>* | No |
ExAC TOPMed gnomAD |
|
| rs367804234 | 1663 | E>G | No |
ESP ExAC TOPMed gnomAD |
|
| rs763241920 | 1663 | E>K | No |
ExAC TOPMed gnomAD |
|
| rs2065719635 | 1664 | Q>H | No | Ensembl | |
| TCGA novel | 1666 | E>K | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065719344 | 1668 | H>Y | No | gnomAD | |
| rs1176561848 | 1669 | N>D | No | gnomAD | |
| rs200372648 | 1670 | I>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs1326950017 | 1670 | I>V | No | TOPMed | |
| rs962147918 | 1672 | D>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
Ensembl NCI-TCGA |
| rs2147325981 | 1673 | E>A | No | Ensembl | |
| rs781512674 | 1675 | T>P | No | ExAC | |
| rs1201371323 | 1678 | I>V | No |
TOPMed gnomAD |
|
| rs867242805 | 1681 | T>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1683 | S>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs376134467 | 1685 | P>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ESP ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 1685 | P>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| COSM5808801 | 1687 | R>Q | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
|
COSM1417059 rs1293031275 |
1687 | R>W | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA gnomAD |
|
COSM4396922 rs1219530033 |
1688 | F>L | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic gnomAD |
| rs369909705 | 1691 | N>S | No |
ESP ExAC TOPMed gnomAD |
|
| rs1428590987 | 1693 | L>F | No |
TOPMed gnomAD |
|
| rs1271536295 | 1697 | H>Q | No |
TOPMed gnomAD |
|
| rs747941915 | 1699 | I>V | No |
ExAC gnomAD |
|
| rs375503886 | 1701 | D>E | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs2147322796 | 1702 | V>M | No | Ensembl | |
| rs751428804 | 1703 | H>N | No |
ExAC gnomAD |
|
| TCGA novel | 1703 | H>Y | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs886094009 | 1704 | V>I | No | TOPMed | |
|
rs1433328187 TCGA novel |
1706 | E>* | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No |
NCI-TCGA gnomAD |
| rs1433328187 | 1706 | E>K | No | gnomAD | |
| rs1193468437 | 1708 | V>A | No | gnomAD | |
| rs2065699694 | 1708 | V>M | No | Ensembl | |
| rs1272970809 | 1710 | A>D | No |
TOPMed gnomAD |
|
| rs1272970809 | 1710 | A>V | No |
TOPMed gnomAD |
|
| rs1199622252 | 1711 | S>L | No |
TOPMed gnomAD |
|
| rs764267512 | 1713 | S>L | No |
ExAC gnomAD |
|
| rs1300156762 | 1718 | T>N | No |
TOPMed gnomAD |
|
| rs1300156762 | 1718 | T>S | No |
TOPMed gnomAD |
|
| rs2065697435 | 1720 | M>L | No | TOPMed | |
| rs780142749 | 1724 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1466941704 | 1725 | R>C | No | gnomAD | |
| rs772755944 | 1725 | R>H | No |
ExAC gnomAD |
|
| rs769389364 | 1726 | T>A | No |
ExAC TOPMed gnomAD |
|
| rs201182504 | 1726 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs1465673046 | 1728 | H>Y | No | Ensembl | |
| rs768379764 | 1732 | R>H | No |
ExAC TOPMed gnomAD |
|
| COSM125457 | 1734 | S>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1601670501 | 1734 | S>P | No | Ensembl | |
| rs1158354478 | 1736 | S>N | No | gnomAD | |
| rs1569155486 | 1738 | K>E | No | Ensembl | |
| TCGA novel | 1739 | L>R | Variant assessed as Somatic; HIGH impact. [NCI-TCGA] | No | NCI-TCGA |
| rs1218834335 | 1746 | S>F | No | gnomAD | |
| rs752886171 | 1747 | T>I | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA gnomAD |
| TCGA novel | 1749 | K>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs929270769 | 1753 | E>K | No | TOPMed | |
| rs763593218 | 1754 | D>E | No |
ExAC gnomAD |
|
| rs201613066 | 1754 | D>G | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs769993436 | 1754 | D>N | No |
ExAC TOPMed gnomAD |
|
| TCGA novel | 1756 | Y>C | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs2065623304 | 1757 | K>E | No | TOPMed | |
| rs775068552 | 1757 | K>R | No |
ExAC TOPMed gnomAD |
|
| rs887763291 | 1758 | G>R | No |
TOPMed gnomAD |
|
| rs770951350 | 1760 | R>L | No |
ExAC TOPMed gnomAD |
|
| rs770951350 | 1760 | R>Q | No |
ExAC TOPMed gnomAD |
|
| rs759338193 | 1760 | R>W | No |
ExAC gnomAD |
|
| rs1330311129 | 1761 | Q>R | No | TOPMed | |
| rs1440319925 | 1769 | D>E | No |
TOPMed gnomAD |
|
| rs542604025 | 1772 | T>A | No |
1000Genomes ExAC TOPMed gnomAD |
|
| rs1334927536 | 1772 | T>R | No | gnomAD | |
| rs1268844021 | 1773 | H>Q | No | Ensembl | |
| rs1047747817 | 1773 | H>R | No | TOPMed | |
| TCGA novel | 1774 | L>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs746289473 | 1775 | A>E | No |
ExAC TOPMed gnomAD |
|
| rs1172981267 | 1776 | E>D | No | gnomAD | |
| rs372200489 | 1779 | R>W | No |
ESP ExAC TOPMed gnomAD |
|
|
COSM1035400 rs372835571 |
1780 | A>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic NCI-TCGA |
| rs1309378692 | 1780 | A>V | No |
TOPMed gnomAD |
|
| rs180698014 | 1781 | H>D | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs180698014 | 1781 | H>Y | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs779890038 | 1782 | T>M | No |
ExAC TOPMed gnomAD |
|
| rs779890038 | 1782 | T>R | No |
ExAC TOPMed gnomAD |
|
| rs750299230 | 1783 | D>E | No |
ExAC gnomAD |
|
| rs376188298 | 1784 | S>F | No |
ESP ExAC TOPMed gnomAD |
|
| rs376188298 | 1784 | S>Y | No |
ESP ExAC TOPMed gnomAD |
|
| rs1375725610 | 1788 | L>F | No | TOPMed | |
| rs1370869646 | 1788 | L>H | No | gnomAD | |
|
rs763150093 COSM3390302 |
1789 | V>M | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA gnomAD |
| rs1254116165 | 1791 | L>F | No | gnomAD | |
| COSM4105089 | 1791 | L>P | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs748709582 | 1797 | Y>C | No |
1000Genomes ExAC gnomAD |
|
| rs2065520014 | 1797 | Y>H | No | Ensembl | |
| rs373532871 | 1798 | T>M | No |
ESP ExAC TOPMed gnomAD |
|
| rs2065519712 | 1798 | T>P | No | Ensembl | |
| rs746546029 | 1799 | Q>R | No |
ExAC gnomAD |
|
|
COSM4105088 rs779693958 COSM4105087 |
1801 | Y>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Cosmic ExAC NCI-TCGA TOPMed gnomAD |
| rs778866905 | 1803 | D>E | No |
ExAC TOPMed gnomAD |
|
| rs1569154158 | 1804 | E>D | No |
TOPMed gnomAD |
|
| rs149124212 | 1804 | E>K | No |
1000Genomes ESP ExAC TOPMed gnomAD |
|
| rs1348330492 | 1805 | I>= | Variant assessed as Somatic; LOW impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 1805 | I>N | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
|
TCGA novel rs2065505648 |
1806 | I>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
NCI-TCGA Ensembl |
|
COSM1566390 COSM4782503 |
1809 | L>F | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs759161065 | 1811 | E>D | No |
ExAC TOPMed gnomAD |
|
| rs1569153963 | 1811 | E>K | No | TOPMed | |
| rs1460120176 | 1812 | D>N | No |
TOPMed gnomAD |
|
| TCGA novel | 1814 | A>V | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs748967251 | 1815 | A>S | No |
ExAC TOPMed gnomAD |
|
| rs748967251 | 1815 | A>T | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No |
ExAC NCI-TCGA TOPMed gnomAD |
| rs777436314 | 1815 | A>V | No |
ExAC gnomAD |
|
| rs1319632932 | 1818 | M>R | No | gnomAD | |
| rs769581020 | 1819 | Q>E | No |
ExAC gnomAD |
|
| rs1383318844 | 1819 | Q>H | No |
TOPMed gnomAD |
|
| rs2065503091 | 1821 | A>S | No | TOPMed | |
| rs1168942832 | 1821 | A>V | No |
TOPMed gnomAD |
|
| rs1022211560 | 1822 | F>L | No |
TOPMed gnomAD |
|
| rs1420220256 | 1822 | F>Y | No | gnomAD | |
|
COSM4810844 COSM1308289 |
1826 | Q>H | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA Cosmic |
| rs1472547858 | 1827 | I>V | No |
TOPMed gnomAD |
|
| rs866516387 | 1828 | A>V | No | Ensembl | |
| TCGA novel | 1829 | A>D | Variant assessed as Somatic; MODERATE impact. [NCI-TCGA] | No | NCI-TCGA |
| rs750555906 | 1829 | A>T | No |
ExAC gnomAD |
|
| rs1445164904 | 1830 | A>T | No |
TOPMed gnomAD |
|
| rs1363450719 | 1830 | A>V | No |
TOPMed gnomAD |
|
| rs2065500450 | 1831 | L>V | No | TOPMed | |
| rs2147287143 | 1834 | K>E | No | Ensembl | |
| rs754031063 | 1835 | V>I | No |
ExAC TOPMed gnomAD |
|
| rs991382066 | 1836 | T>A | No | Ensembl | |
| rs2065499062 | 1836 | T>I | No | Ensembl |
No associated diseases with O15031
12 regional properties for O15031
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Sema domain | 8 - 466 | IPR001627 |
| repeat | Plexin repeat | 469 - 518 | IPR002165-1 |
| repeat | Plexin repeat | 759 - 798 | IPR002165-2 |
| domain | IPT domain | 802 - 893 | IPR002909-1 |
| domain | IPT domain | 894 - 980 | IPR002909-2 |
| domain | IPT domain | 982 - 1092 | IPR002909-3 |
| domain | Plexin, cytoplasmic RasGAP domain | 1271 - 1805 | IPR013548 |
| domain | PSI domain | 468 - 519 | IPR016201-1 |
| domain | PSI domain | 614 - 667 | IPR016201-2 |
| domain | PSI domain | 759 - 801 | IPR016201-3 |
| domain | Plexin, TIG domain 1 | 525 - 613 | IPR041019 |
| domain | Plexin, cytoplasmic RhoGTPase-binding domain | 1448 - 1556 | IPR046800 |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell surface | The external part of the cell wall and/or plasma membrane. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| semaphorin receptor complex | A stable binary complex of a neurophilin and a plexin, together forming a functional semaphorin receptor. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| semaphorin receptor activity | Combining with a semaphorin, and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity. |
16 GO annotations of biological process
| Name | Definition |
|---|---|
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| excitatory synapse assembly | The aggregation, arrangement and bonding together of a set of components to form an excitatory synapse. |
| homophilic cell adhesion via plasma membrane adhesion molecules | The attachment of a plasma membrane adhesion molecule in one cell to an identical molecule in an adjacent cell. |
| negative regulation of cell adhesion | Any process that stops, prevents, or reduces the frequency, rate or extent of cell adhesion. |
| neural tube closure | The last step in the formation of the neural tube, where the paired neural folds are brought together and fuse at the dorsal midline. |
| neuroblast proliferation | The expansion of a neuroblast population by cell division. A neuroblast is any cell that will divide and give rise to a neuron. |
| positive regulation of axonogenesis | Any process that activates or increases the frequency, rate or extent of axonogenesis. |
| positive regulation of neuron projection development | Any process that increases the rate, frequency or extent of neuron projection development. Neuron projection development is the process whose specific outcome is the progression of a neuron projection over time, from its formation to the mature structure. A neuron projection is any process extending from a neural cell, such as axons or dendrites (collectively called neurites). |
| positive regulation of translation | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| regulation of cell migration | Any process that modulates the frequency, rate or extent of cell migration. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of GTPase activity | Any process that modulates the rate of GTP hydrolysis by a GTPase. |
| regulation of neuron migration | Any process that modulates the frequency, rate or extent of neuron migration. |
| regulation of protein phosphorylation | Any process that modulates the frequency, rate or extent of addition of phosphate groups into an amino acid in a protein. |
| semaphorin-plexin signaling pathway | The series of molecular signals generated as a consequence of a semaphorin receptor (composed of a plexin and a neurophilin) binding to a semaphorin ligand. |
| semaphorin-plexin signaling pathway involved in axon guidance | Any semaphorin-plexin signaling pathway that is involved in axon guidance. |
27 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q769I5 | MET | Hepatocyte growth factor receptor | Bos taurus (Bovine) | PR |
| A0M8S8 | MET | Hepatocyte growth factor receptor | Felis catus (Cat) (Felis silvestris catus) | PR |
| Q04912 | MST1R | Macrophage-stimulating protein receptor | Homo sapiens (Human) | EV |
| P08581 | MET | Hepatocyte growth factor receptor | Homo sapiens (Human) | EV |
| P51805 | PLXNA3 | Plexin-A3 | Homo sapiens (Human) | SS |
| Q9UIW2 | PLXNA1 | Plexin-A1 | Homo sapiens (Human) | EV SS |
| Q9ULL4 | PLXNB3 | Plexin-B3 | Homo sapiens (Human) | SS |
| O75051 | PLXNA2 | Plexin-A2 | Homo sapiens (Human) | SS |
| O43157 | PLXNB1 | Plexin-B1 | Homo sapiens (Human) | EV SS |
| Q9HCM2 | PLXNA4 | Plexin-A4 | Homo sapiens (Human) | SS |
| Q9QY40 | Plxnb3 | Plexin-B3 | Mus musculus (Mouse) | SS |
| P70208 | Plxna3 | Plexin-A3 | Mus musculus (Mouse) | EV SS |
| Q3UH93 | Plxnd1 | Plexin-D1 | Mus musculus (Mouse) | SS |
| P70207 | Plxna2 | Plexin-A2 | Mus musculus (Mouse) | SS |
| Q62190 | Mst1r | Macrophage-stimulating protein receptor | Mus musculus (Mouse) | SS |
| Q8CJH3 | Plxnb1 | Plexin-B1 | Mus musculus (Mouse) | SS |
| Q9QZC2 | Plxnc1 | Plexin-C1 | Mus musculus (Mouse) | SS |
| Q80UG2 | Plxna4 | Plexin-A4 | Mus musculus (Mouse) | SS |
| P16056 | Met | Hepatocyte growth factor receptor | Mus musculus (Mouse) | PR |
| P70206 | Plxna1 | Plexin-A1 | Mus musculus (Mouse) | EV SS |
| B2RXS4 | Plxnb2 | Plexin-B2 | Mus musculus (Mouse) | SS |
| Q2QLE0 | MET | Hepatocyte growth factor receptor | Sus scrofa (Pig) | PR |
| P97523 | Met | Hepatocyte growth factor receptor | Rattus norvegicus (Rat) | PR |
| D3ZPX4 | Plxna3 | Plexin-A3 | Rattus norvegicus (Rat) | SS |
| D3ZLH5 | Plxnb3 | Plexin-B3 | Rattus norvegicus (Rat) | SS |
| Q6BEA0 | plxna4 | Plexin-A4 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| B0S5N4 | plxna3 | Plexin A3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MALQLWALTL | LGLLGAGASL | RPRKLDFFRS | EKELNHLAVD | EASGVVYLGA | VNALYQLDAK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LQLEQQVATG | PALDNKKCTP | PIEASQCHEA | EMTDNVNQLL | LLDPPRKRLV | ECGSLFKGIC |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ALRALSNISL | RLFYEDGSGE | KSFVASNDEG | VATVGLVSST | GPGGDRVLFV | GKGNGPHDNG |
| 190 | 200 | 210 | 220 | 230 | 240 |
| IIVSTRLLDR | TDSREAFEAY | TDHATYKAGY | LSTNTQQFVA | AFEDGPYVFF | VFNQQDKHPA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RNRTLLARMC | REDPNYYSYL | EMDLQCRDPD | IHAAAFGTCL | AASVAAPGSG | RVLYAVFSRD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SRSSGGPGAG | LCLFPLDKVH | AKMEANRNAC | YTGTREARDI | FYKPFHGDIQ | CGGHAPGSSK |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SFPCGSEHLP | YPLGSRDGLR | GTAVLQRGGL | NLTAVTVAAE | NNHTVAFLGT | SDGRILKVYL |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TPDGTSSEYD | SILVEINKRV | KRDLVLSGDL | GSLYAMTQDK | VFRLPVQECL | SYPTCTQCRD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SQDPYCGWCV | VEGRCTRKAE | CPRAEEASHW | LWSRSKSCVA | VTSAQPQNMS | RRAQGEVQLT |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VSPLPALSEE | DELLCLFGES | PPHPARVEGE | AVICNSPSSI | PVTPPGQDHV | AVTIQLLLRR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| GNIFLTSYQY | PFYDCRQAMS | LEENLPCISC | VSNRWTCQWD | LRYHECREAS | PNPEDGIVRA |
| 670 | 680 | 690 | 700 | 710 | 720 |
| HMEDSCPQFL | GPSPLVIPMN | HETDVNFQGK | NLDTVKGSSL | HVGSDLLKFM | EPVTMQESGT |
| 730 | 740 | 750 | 760 | 770 | 780 |
| FAFRTPKLSH | DANETLPLHL | YVKSYGKNID | SKLHVTLYNC | SFGRSDCSLC | RAANPDYRCA |
| 790 | 800 | 810 | 820 | 830 | 840 |
| WCGGQSRCVY | EALCNTTSEC | PPPVITRIQP | ETGPLGGGIR | ITILGSNLGV | QAGDIQRISV |
| 850 | 860 | 870 | 880 | 890 | 900 |
| AGRNCSFQPE | RYSVSTRIVC | VIEAAETPFT | GGVEVDVFGK | LGRSPPNVQF | TFQQPKPLSV |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EPQQGPQAGG | TTLTIHGTHL | DTGSQEDVRV | TLNGVPCKVT | KFGAQLQCVT | GPQATRGQML |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| LEVSYGGSPV | PNPGIFFTYR | ENPVLRAFEP | LRSFASGGRS | INVTGQGFSL | IQRFAMVVIA |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| EPLQSWQPPR | EAESLQPMTV | VGTDYVFHND | TKVVFLSPAV | PEEPEAYNLT | VLIEMDGHRA |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| LLRTEAGAFE | YVPDPTFENF | TGGVKKQVNK | LIHARGTNLN | KAMTLQEAEA | FVGAERCTMK |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| TLTETDLYCE | PPEVQPPPKR | RQKRDTTHNL | PEFIVKFGSR | EWVLGRVEYD | TRVSDVPLSL |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| ILPLVIVPMV | VVIAVSVYCY | WRKSQQAERE | YEKIKSQLEG | LEESVRDRCK | KEFTDLMIEM |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| EDQTNDVHEA | GIPVLDYKTY | TDRVFFLPSK | DGDKDVMITG | KLDIPEPRRP | VVEQALYQFS |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| NLLNSKSFLI | NFIHTLENQR | EFSARAKVYF | ASLLTVALHG | KLEYYTDIMH | TLFLELLEQY |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| VVAKNPKLML | RRSETVVERM | LSNWMSICLY | QYLKDSAGEP | LYKLFKAIKH | QVEKGPVDAV |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| QKKAKYTLND | TGLLGDDVEY | APLTVSVIVQ | DEGVDAIPVK | VLNCDTISQV | KEKIIDQVYR |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| GQPCSCWPRP | DSVVLEWRPG | STAQILSDLD | LTSQREGRWK | RVNTLMHYNV | RDGATLILSK |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| VGVSQQPEDS | QQDLPGERHA | LLEEENRVWH | LVRPTDEVDE | GKSKRGSVKE | KERTKAITEI |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| YLTRLLSVKG | TLQQFVDNFF | QSVLAPGHAV | PPAVKYFFDF | LDEQAEKHNI | QDEDTIHIWK |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| TNSLPLRFWV | NILKNPHFIF | DVHVHEVVDA | SLSVIAQTFM | DACTRTEHKL | SRDSPSNKLL |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| YAKEISTYKK | MVEDYYKGIR | QMVQVSDQDM | NTHLAEISRA | HTDSLNTLVA | LHQLYQYTQK |
| 1810 | 1820 | 1830 | |||
| YYDEIINALE | EDPAAQKMQL | AFRLQQIAAA | LENKVTDL |