O14770
Gene name |
MEIS2 (MRG1) |
Protein name |
Homeobox protein Meis2 |
Names |
Meis1-related protein 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4212 |
EC number |
|
Protein Class |
HOMEOBOX PROTEIN TRANSCRIPTION FACTORS (PTHR11850) |
Descriptions
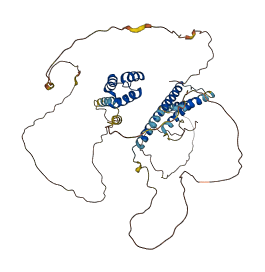
MEIS2 is a homeodomain protein containing a conserved homothorax (Hth) domain that is present in all Meis and Prep family proteins and in the Drosophila Hth protein. The region (150-193 residues) of MEIS2 is an autoinhibitory region which encompasses homology region 2 (hr2) of the Hth domain. The Hth domain interacts with Pbx proteins, thereby promoting cooperative binding of Meis/Pbx dimers to a composite DNA element, and this interaction facilitates binding of the Pbx partner to DNA.
Autoinhibitory domains (AIDs)
Target domain |
340-470 (C-terminal transcriptional activation domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

5 structures for O14770
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 3K2A | X-ray | 195 A | A/B | 281-345 | PDB |
| 4XRM | X-ray | 160 A | A/B | 281-342 | PDB |
| 5BNG | X-ray | 350 A | A/B | 283-342 | PDB |
| 5EG0 | X-ray | 310 A | A | 284-338 | PDB |
| AF-O14770-F1 | Predicted | AlphaFoldDB |
269 variants for O14770
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV001335516 rs372646327 |
56 | P>Q | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000490249 rs749346955 CA391927613 |
204 | S>* | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
RCV000677393 rs1555456994 |
276 | R>missing | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001335517 rs1385058993 |
293 | A>P | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies [ClinVar] | Yes |
ClinVar dbSNP |
|
CA16619921 rs1064793383 RCV002221236 RCV000482887 |
302 | P>L | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000989283 rs1595790647 RCV001265871 |
312 | L>missing | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2058712732 RCV001252071 |
322 | Q>E | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000509419 RCV001267141 rs879255264 RCV000210625 RCV000494204 |
333 | R>missing | Cardiac malformation, cleft lip/palate, microcephaly, and digital anomalies MEIS2-related disorder Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
CA391929335 rs1226507814 |
4 | R>G | No |
ClinGen gnomAD |
|
|
CA391929331 rs1381339489 |
4 | R>M | No |
ClinGen gnomAD |
|
|
CA7469501 rs745617695 |
5 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748786042 CA391929302 |
6 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391929303 rs748786042 |
6 | D>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757192671 CA7469499 |
6 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs778738819 CA7469500 |
6 | D>H | No |
ClinGen ExAC gnomAD |
|
|
CA391929304 rs757192671 |
6 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777329350 CA7469497 |
7 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391929291 rs1229352147 |
8 | L>P | No |
ClinGen TOPMed gnomAD |
|
|
CA391929290 rs1229352147 |
8 | L>R | No |
ClinGen TOPMed gnomAD |
|
|
CA7469494 rs767120205 |
10 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA7469495 rs767120205 |
10 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA269148374 rs957652925 |
12 | G>R | No |
ClinGen gnomAD |
|
|
rs1555473800 RCV000497572 CA391929261 |
13 | G>E | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs937865015 CA269148373 |
14 | M>V | No |
ClinGen Ensembl |
|
|
rs1283836405 CA391929237 |
16 | G>E | No |
ClinGen TOPMed |
|
|
CA269148372 rs927806018 |
17 | V>G | No |
ClinGen Ensembl |
|
|
CA269148371 rs865880036 |
19 | V>G | No |
ClinGen gnomAD |
|
|
CA7469490 rs762378662 |
19 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA7469488 rs764284757 |
21 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA391929201 rs1031517719 |
23 | M>K | No |
ClinGen TOPMed gnomAD |
|
|
CA391929200 rs1031517719 |
23 | M>R | No |
ClinGen TOPMed gnomAD |
|
|
CA269148370 rs1031517719 |
23 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
CA7469487 rs760768956 |
25 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1253953785 CA391929180 |
26 | D>A | No |
ClinGen gnomAD |
|
|
rs1253953785 CA391929179 |
26 | D>G | No |
ClinGen gnomAD |
|
|
CA391929175 rs1482810038 |
27 | P>T | No |
ClinGen gnomAD |
|
|
rs1272742788 CA391929162 |
28 | H>Q | No |
ClinGen gnomAD |
|
|
COSM1152821 rs772370515 CA7469485 |
29 | A>V | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA391929151 rs1259994950 |
30 | P>L | No |
ClinGen TOPMed |
|
|
CA391929154 rs1212253316 |
30 | P>S | No |
ClinGen TOPMed |
|
|
CA391929139 rs774160502 |
33 | I>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391929134 rs749233162 |
33 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774160502 CA7469483 |
33 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1289568511 CA391929131 COSM1517057 COSM1517058 |
34 | P>S | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA391929123 rs1274403454 |
35 | P>L | No |
ClinGen gnomAD |
|
|
rs138503850 CA7469479 |
35 | P>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1039255246 CA269148369 |
36 | V>I | No |
ClinGen TOPMed |
|
|
CA7469476 rs754593480 |
37 | H>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469477 rs754593480 |
37 | H>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469475 rs750820203 |
37 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
rs144628203 CA7469474 |
40 | N>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1316493017 CA391929082 |
42 | G>R | No |
ClinGen gnomAD |
|
|
rs754372405 CA7469472 |
44 | P>Q | No |
ClinGen ExAC gnomAD |
|
|
CA391929054 rs760859088 |
46 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391929056 rs1472270409 |
46 | H>R | No |
ClinGen gnomAD |
|
|
rs1596139159 CA391929045 |
48 | T>A | No |
ClinGen Ensembl |
|
|
rs918913476 CA269148368 |
52 | G>R | No |
ClinGen TOPMed |
|
|
CA391929015 rs918913476 |
52 | G>S | No |
ClinGen TOPMed |
|
|
CA7469463 rs773159665 |
55 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs372646327 COSM1587816 CA7469460 COSM960957 |
56 | P>L | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs372646327 CA7469461 |
56 | P>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1379402160 CA391928976 |
58 | P>H | No |
ClinGen gnomAD |
|
|
rs1397317991 CA391928979 |
58 | P>T | No |
ClinGen gnomAD |
|
|
CA391928967 rs1438556989 |
59 | N>K | No |
ClinGen TOPMed |
|
|
rs780677066 CA7469459 |
59 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA269148366 rs768355698 |
61 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469458 rs768355698 |
61 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469457 rs746689909 |
62 | P>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391928950 rs746689909 |
62 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1379369473 CA391928951 |
62 | P>S | No |
ClinGen TOPMed |
|
|
rs926831373 CA269148365 |
64 | S>G | No |
ClinGen TOPMed |
|
|
rs1456673066 CA391928929 |
65 | M>I | No |
ClinGen gnomAD |
|
|
CA7469455 rs757669617 |
67 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA391928914 rs1424515511 |
68 | A>T | No |
ClinGen gnomAD |
|
|
rs1255733126 CA391928910 |
68 | A>V | No |
ClinGen gnomAD |
|
|
CA391928907 rs1449220711 |
69 | V>F | No |
ClinGen TOPMed |
|
|
rs1466484335 CA391928874 |
74 | K>Q | No |
ClinGen gnomAD |
|
|
rs1208597242 CA391928832 COSM321662 |
79 | A>V | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed gnomAD |
|
CA391928767 rs1387283814 |
87 | P>S | No |
ClinGen gnomAD |
|
|
rs763081606 CA7469423 |
90 | A>S | No |
ClinGen ExAC gnomAD |
|
|
CA391928697 rs1344639476 |
97 | E>D | No |
ClinGen TOPMed |
|
|
CA391928656 rs1400775761 |
104 | R>Q | No |
ClinGen TOPMed |
|
|
CA391928632 rs1474596915 |
108 | V>M | No |
ClinGen gnomAD |
|
|
CA391928601 COSM78075 rs1214102280 COSM1587818 |
113 | V>I | ovary endometrium [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
CA391928574 rs776583289 |
117 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs776583289 CA7469419 |
117 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1323988867 CA391928524 |
123 | I>M | No |
ClinGen TOPMed |
|
|
rs1220911568 CA391928513 |
125 | V>A | No |
ClinGen TOPMed |
|
|
rs760278105 CA7469417 |
129 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA269148099 rs200496429 |
134 | K>E | No |
ClinGen 1000Genomes |
|
|
CA391928426 rs1322616497 |
136 | L>P | No |
ClinGen gnomAD |
|
|
rs760522172 CA7469399 |
138 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA391928389 rs1567294240 RCV000760827 |
142 | E>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA391928191 rs1156581518 |
162 | E>G | No |
ClinGen TOPMed |
|
|
rs1473337121 CA391928194 |
162 | E>Q | No |
ClinGen TOPMed |
|
|
CA269147913 rs1039511168 |
169 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
CA391927966 rs1555471981 |
174 | R>* | No |
ClinGen Ensembl |
|
|
CA7469358 rs752485891 |
174 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA7469357 rs368930165 |
176 | I>V | No |
ClinGen ESP ExAC gnomAD |
|
|
COSM2268535 COSM2268534 CA391927828 rs1263005930 |
186 | D>N | large_intestine [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs762490597 CA7469353 |
188 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391927805 rs762490597 |
188 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1322337654 CA391927747 |
192 | R>S | No |
ClinGen TOPMed |
|
|
CA391927744 rs1365751131 |
193 | D>H | No |
ClinGen TOPMed |
|
|
rs1230534167 CA391927738 |
193 | D>V | No |
ClinGen TOPMed |
|
|
rs571091781 CA7469350 |
194 | G>C | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs571091781 CA7469351 |
194 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7469349 rs775890981 |
194 | G>V | No |
ClinGen ExAC gnomAD |
|
|
CA7469347 rs760019942 |
197 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1449882838 CA391927656 |
200 | H>Q | No |
ClinGen gnomAD |
|
|
rs774778254 CA7469346 |
202 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA7469345 rs185278462 |
203 | L>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7469344 rs749346955 |
204 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1264692993 CA391927604 |
205 | G>A | No |
ClinGen gnomAD |
|
|
rs770033702 CA7469342 |
206 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391927601 rs1159880021 |
206 | S>P | No |
ClinGen gnomAD |
|
|
rs1248730273 CA391927579 |
208 | T>A | No |
ClinGen TOPMed |
|
|
rs566632679 CA7469338 |
211 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs548323279 CA7469337 |
212 | D>E | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA391927455 rs1310960333 |
214 | N>I | No |
ClinGen TOPMed |
|
|
rs1258457335 CA391927452 |
215 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
CA7469319 rs758327782 |
215 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1567275949 CA391927422 |
219 | R>Q | No |
ClinGen Ensembl |
|
|
rs1330564715 CA391927418 |
220 | D>A | No |
ClinGen TOPMed |
|
|
rs753490361 COSM1214904 CA7469315 COSM1214903 |
222 | D>N | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA391927391 rs1329224401 |
223 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA391927381 rs1596101188 |
225 | T>S | No |
ClinGen Ensembl |
|
|
rs1359901644 CA391927371 |
227 | T>A | No |
ClinGen gnomAD |
|
|
CA391927367 rs1295342702 |
227 | T>I | No |
ClinGen gnomAD |
|
|
CA7469313 rs147874392 |
228 | H>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA391927342 rs1255085132 |
231 | G>V | No |
ClinGen TOPMed |
|
|
CA7469310 rs763504614 |
235 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA7469309 rs773535694 |
238 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs1892547180 RCV001090729 |
239 | G>missing | No |
ClinVar dbSNP |
|
|
CA7469307 rs761760302 |
240 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA7469308 rs765363630 |
240 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA391927263 rs1200949253 |
244 | S>N | No |
ClinGen gnomAD |
|
|
CA7469305 rs375357305 |
245 | G>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs376747133 CA7469304 |
248 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391927230 rs1251699935 |
249 | S>G | No |
ClinGen gnomAD |
|
|
rs370758497 CA391927216 |
250 | E>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA269141780 rs1048086762 |
254 | G>C | No |
ClinGen TOPMed gnomAD |
|
|
CA7469278 rs777260011 |
254 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1446092603 CA391928136 |
257 | N>D | No |
ClinGen gnomAD |
|
|
CA7469277 rs769237139 |
258 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1435798806 CA391928119 |
259 | V>A | No |
ClinGen gnomAD |
|
|
rs780669823 CA7469275 |
262 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391928070 rs1186393341 |
263 | G>A | No |
ClinGen gnomAD |
|
|
rs779516436 CA7469272 |
269 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs754026070 CA7469270 |
270 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs369609025 CA7469269 |
270 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs752990884 CA7469267 |
271 | D>Y | No |
ClinGen ExAC TOPMed |
|
|
CA391927872 rs1406144240 |
275 | K>I | No |
ClinGen TOPMed |
|
|
CA391927859 rs1452423816 |
276 | R>C | No |
ClinGen gnomAD |
|
|
rs767585793 COSM1562967 CA7469266 COSM1562966 |
276 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
COSM224734 CA269141779 rs267604162 |
284 | P>S | skin [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA391927624 rs1401881711 |
290 | I>V | No |
ClinGen gnomAD |
|
|
rs1385058993 CA391927569 |
293 | A>S | No |
ClinGen TOPMed gnomAD |
|
|
CA391927572 rs1385058993 |
293 | A>T | No |
ClinGen TOPMed gnomAD |
|
|
CA391927096 rs1275580589 |
313 | A>V | No |
ClinGen gnomAD |
|
|
CA391927058 rs1567094842 |
319 | T>K | No |
ClinGen Ensembl |
|
|
RCV000487166 rs1064796723 CA16619920 |
322 | Q>L | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1131691404 RCV000494298 CA391926969 |
329 | N>K | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA391926899 rs1372284638 |
339 | I>T | No |
ClinGen gnomAD |
|
|
rs1327678536 CA391926826 |
348 | L>P | No |
ClinGen gnomAD |
|
|
CA7469195 rs368127920 |
349 | L>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1172909836 CA391926787 |
354 | S>N | No |
ClinGen TOPMed |
|
|
CA391926767 rs1165717358 |
357 | A>T | No |
ClinGen gnomAD |
|
|
rs759249181 CA7469193 |
358 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1385043709 CA391926748 |
360 | S>G | No |
ClinGen gnomAD |
|
|
rs1430791975 CA391926689 |
368 | S>N | No |
ClinGen gnomAD |
|
|
CA391926684 rs1442100305 |
369 | F>I | No |
ClinGen TOPMed |
|
|
rs1433862229 CA391926675 |
370 | V>L | No |
ClinGen gnomAD |
|
|
rs1186578130 CA391926658 |
372 | D>E | No |
ClinGen gnomAD |
|
|
rs1027874815 CA391926663 |
372 | D>H | No |
ClinGen TOPMed |
|
|
rs1027874815 CA269126524 |
372 | D>Y | No |
ClinGen TOPMed |
|
|
rs1047475739 CA269126523 |
373 | G>S | No |
ClinGen gnomAD |
|
|
rs534636474 CA7469191 |
377 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs534636474 CA7469190 |
377 | M>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391926627 rs534636474 |
377 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391926604 rs1595666758 |
380 | R>Q | No |
ClinGen Ensembl |
|
|
CA269126522 rs895441531 |
381 | P>S | No |
ClinGen TOPMed |
|
|
CA7469148 rs775995092 |
384 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469147 rs767985608 |
386 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA7469146 rs759650078 |
386 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774482745 CA7469145 |
387 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs531139958 CA7469144 |
389 | G>A | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA391926386 rs531139958 |
389 | G>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1447970871 CA391926377 |
390 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1339277809 CA391926372 |
391 | Y>C | No |
ClinGen gnomAD |
|
|
CA7469142 rs151055354 |
392 | V>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7469141 rs142878520 |
394 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA269126261 rs987970014 |
395 | G>V | No |
ClinGen TOPMed |
|
|
rs560301639 CA7469140 |
396 | G>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs930989017 CA269126260 |
398 | M>T | No |
ClinGen Ensembl |
|
|
CA7469139 rs781319540 |
398 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA391926327 rs1386881633 |
399 | G>R | No |
ClinGen gnomAD |
|
|
rs754963373 CA7469138 |
400 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs1338459502 CA391926283 |
404 | Q>H | No |
ClinGen gnomAD |
|
|
CA269126259 rs113747758 |
405 | P>L | No |
ClinGen Ensembl |
|
|
rs956554759 CA269126258 |
406 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1195398289 CA391926249 |
410 | P>T | No |
ClinGen gnomAD |
|
|
CA391926235 rs1447667587 |
412 | M>V | No |
ClinGen gnomAD |
|
|
CA7469137 rs746560276 |
413 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs1214763170 CA391926220 |
414 | P>S | No |
ClinGen gnomAD |
|
|
CA269126256 rs1032019811 |
415 | H>L | No |
ClinGen TOPMed |
|
|
CA391926214 rs1032019811 |
415 | H>P | No |
ClinGen TOPMed |
|
|
CA391926212 rs1292932507 |
415 | H>Q | No |
ClinGen gnomAD |
|
|
rs779632587 CA7469136 |
415 | H>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs757783328 CA7469135 |
418 | Q>P | No |
ClinGen ExAC gnomAD |
|
|
rs144548752 CA7469133 |
421 | H>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA391926165 rs1340627805 |
422 | G>V | No |
ClinGen gnomAD |
|
|
rs768077389 CA7469130 |
423 | P>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391926163 rs753026080 |
423 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs753026080 CA7469131 |
423 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs1358602257 CA391926158 |
424 | P>S | No |
ClinGen TOPMed |
|
|
CA7469129 rs760021973 |
425 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1404611090 CA391926141 |
426 | H>L | No |
ClinGen gnomAD |
|
|
rs533167442 CA7469127 |
429 | L>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA7469126 rs762901201 |
430 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs1360412743 CA391926112 |
431 | S>G | No |
ClinGen TOPMed |
|
|
CA391926104 rs1445101744 |
432 | H>Y | No |
ClinGen TOPMed |
|
|
CA391926093 rs1283242514 |
433 | P>R | No |
ClinGen TOPMed |
|
|
CA7469125 rs773532150 |
434 | H>P | No |
ClinGen ExAC |
|
|
rs1191659094 CA391926091 |
434 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs770111561 CA7469124 |
435 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA391926073 rs1269960840 |
436 | P>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA391926070 rs374458760 |
437 | A>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7469123 rs374458760 |
437 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA269126254 rs200282136 |
438 | M>I | No |
ClinGen 1000Genomes gnomAD |
|
|
rs141054432 CA7469122 |
438 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA391926053 rs1201550451 |
439 | M>I | No |
ClinGen TOPMed |
|
|
rs768515170 CA7469121 |
439 | M>T | No |
ClinGen ExAC gnomAD |
|
|
rs746974524 CA7469120 |
440 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA391926032 rs1346990815 |
442 | G>A | No |
ClinGen gnomAD |
|
|
rs201588285 CA7469118 |
442 | G>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA391926034 rs201588285 |
442 | G>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA7469116 rs778573000 |
443 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA391926025 rs1183813879 |
444 | P>T | No |
ClinGen TOPMed gnomAD |
|
|
rs753115894 CA7469114 |
445 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA7469115 rs756895354 |
445 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469112 rs145933742 |
447 | H>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA269126252 rs370135711 |
448 | P>H | No |
ClinGen ESP |
|
|
CA391925981 rs1481495476 |
451 | T>A | No |
ClinGen gnomAD |
|
|
rs142325445 CA7469111 |
451 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs377210457 CA7469109 |
452 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA7469108 rs763133151 |
454 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs372766056 CA7469107 |
455 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA391925932 rs1457704441 |
458 | T>I | No |
ClinGen gnomAD |
|
|
rs765614880 CA7469106 |
459 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762141306 CA7469105 |
463 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA269126250 rs898671361 |
465 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs776929937 CA7469104 |
466 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA7469102 rs760342372 COSM1562968 |
469 | G>R | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA391925854 rs1202750626 |
470 | Q>H | No |
ClinGen Ensembl |
|
|
rs775551671 CA7469101 |
470 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs771868780 CA7469100 |
471 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748942972 CA7469096 |
472 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA7469097 rs150313361 |
472 | M>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs542478499 CA7469098 |
472 | M>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA7469095 rs575475928 |
473 | D>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA7469094 rs575475928 |
473 | D>Y | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA7469093 rs751839152 |
475 | H>R | No |
ClinGen ExAC gnomAD |
|
|
rs369601092 CA7469092 |
476 | A>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA269126249 rs937401157 |
477 | Q>* | No |
ClinGen TOPMed |
|
|
CA391925814 rs758819750 |
477 | Q>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391925815 rs758819750 |
477 | Q>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs758819750 CA7469091 |
477 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA391925807 rs1256052887 |
478 | Q>W | No |
ClinGen gnomAD |
1 associated diseases with O14770
[MIM: 600987]: Cleft palate, cardiac defects, and intellectual disability (CPCMR)
An autosomal dominant disease characterized by multiple congenital malformations, mild-to-severe intellectual disability with poor speech, and delayed psychomotor development. Congenital malformations include heart defects, cleft lip/palate, distally-placed thumbs and toes, and cutaneous syndactyly between the second and third toes. {ECO:0000269|PubMed:24678003, ECO:0000269|PubMed:25712757, ECO:0000269|PubMed:27225850}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- An autosomal dominant disease characterized by multiple congenital malformations, mild-to-severe intellectual disability with poor speech, and delayed psychomotor development. Congenital malformations include heart defects, cleft lip/palate, distally-placed thumbs and toes, and cutaneous syndactyly between the second and third toes. {ECO:0000269|PubMed:24678003, ECO:0000269|PubMed:25712757, ECO:0000269|PubMed:27225850}. Note=The disease is caused by variants affecting the gene represented in this entry.
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11850 | HOMEOBOX PROTEIN TRANSCRIPTION FACTORS |
| PANTHER Subfamily | PTHR11850:SF47 | HOMEOBOX PROTEIN MEIS2 |
| PANTHER Protein Class |
homeodomain transcription factor
DNA-binding transcription factor helix-turn-helix transcription factor |
|
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
| sequence-specific double-stranded DNA binding | Binding to double-stranded DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA, e.g. promotor binding or rDNA binding. |
| transcription factor binding | Binding to a transcription factor, a protein required to initiate or regulate transcription. |
15 GO annotations of biological process
| Name | Definition |
|---|---|
| animal organ morphogenesis | Morphogenesis of an animal organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process in which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| embryonic pattern specification | The process that results in the patterns of cell differentiation that will arise in an embryo. |
| eye development | The process whose specific outcome is the progression of the eye over time, from its formation to the mature structure. The eye is the organ of sight. |
| negative regulation of myeloid cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of myeloid cell differentiation. |
| negative regulation of transcription by RNA polymerase II | Any process that stops, prevents, or reduces the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| pancreas development | The process whose specific outcome is the progression of the pancreas over time, from its formation to the mature structure. The pancreas is an endoderm derived structure that produces precursors of digestive enzymes and blood glucose regulating enzymes. |
| positive regulation of cardiac muscle myoblast proliferation | Any process that activates or increases the frequency, rate or extent of cardiac muscle myoblast proliferation. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of mitotic cell cycle | Any process that activates or increases the rate or extent of progression through the mitotic cell cycle. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
| response to growth factor | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a growth factor stimulus. |
| response to mechanical stimulus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| visual learning | Any process in an organism in which a change in behavior of an individual occurs in response to repeated exposure to a visual cue. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8MIB7 | TGIF2LX | Homeobox protein TGIF2LX | Pan troglodytes (Chimpanzee) | PR |
| P40424 | PBX1 | Pre-B-cell leukemia transcription factor 1 | Homo sapiens (Human) | PR |
| P40425 | PBX2 | Pre-B-cell leukemia transcription factor 2 | Homo sapiens (Human) | PR |
| Q8IUE1 | TGIF2LX | Homeobox protein TGIF2LX | Homo sapiens (Human) | PR |
| Q99687 | MEIS3 | Homeobox protein Meis3 | Homo sapiens (Human) | SS |
| O00470 | MEIS1 | Homeobox protein Meis1 | Homo sapiens (Human) | SS |
| A8K0S8 | MEIS3P2 | Putative homeobox protein Meis3-like 2 | Homo sapiens (Human) | PR |
| A6NDR6 | MEIS3P1 | Putative homeobox protein Meis3-like 1 | Homo sapiens (Human) | PR |
| P97368 | Meis3 | Homeobox protein Meis3 | Mus musculus (Mouse) | PR |
| P97367 | Meis2 | Homeobox protein Meis2 | Mus musculus (Mouse) | SS |
| Q75LX9 | Os03g0673500 | Putative homeobox protein knotted-1-like 5 | Oryza sativa subsp japonica (Rice) | PR |
| Q75LX7 | OSH10 | Homeobox protein knotted-1-like 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q1PFD1 | BLH11 | BEL1-like homeodomain protein 11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIW1 | BLH7 | BEL1-like homeodomain protein 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAQRYDELPH | YGGMDGVGVP | ASMYGDPHAP | RPIPPVHHLN | HGPPLHATQH | YGAHAPHPNV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| MPASMGSAVN | DALKRDKDAI | YGHPLFPLLA | LVFEKCELAT | CTPREPGVAG | GDVCSSDSFN |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EDIAVFAKQV | RAEKPLFSSN | PELDNLMIQA | IQVLRFHLLE | LEKVHELCDN | FCHRYISCLK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GKMPIDLVID | ERDGSSKSDH | EELSGSSTNL | ADHNPSSWRD | HDDATSTHSA | GTPGPSSGGH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ASQSGDNSSE | QGDGLDNSVA | SPGTGDDDDP | DKDKKRQKKR | GIFPKVATNI | MRAWLFQHLT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| HPYPSEEQKK | QLAQDTGLTI | LQVNNWFINA | RRRIVQPMID | QSNRAGFLLD | PSVSQGAAYS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PEGQPMGSFV | LDGQQHMGIR | PAGLQSMPGD | YVSQGGPMGM | SMAQPSYTPP | QMTPHPTQLR |
| 430 | 440 | 450 | 460 | 470 | |
| HGPPMHSYLP | SHPHHPAMMM | HGGPPTHPGM | TMSAQSPTML | NSVDPNVGGQ | VMDIHAQ |