O14757
Gene name |
CHEK1 (CHK1) |
Protein name |
Serine/threonine-protein kinase Chk1 |
Names |
CHK1 checkpoint homolog, Cell cycle checkpoint kinase, Checkpoint kinase-1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1111 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
MAP/MICROTUBULE AFFINITY-REGULATING KINASE (PTHR24346) |
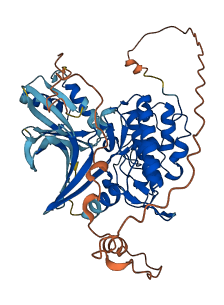
Descriptions
CHEK1 is a serine/threonine protein kinase and is a key mediator that links the DNA stress responses to the cell cycle. Full-length CHEK1 has only low basal activity when expressed. Truncation of the C-terminal regulatory region activates CHEK1 even in the absence of stress.
Autoinhibitory domains (AIDs)
Target domain |
9-265 (Protein kinase domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment |
Accessory elements
147-172 (Activation loop from InterPro)
Target domain |
9-265 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

160 structures for O14757
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1IA8 | X-ray | 170 A | A | 1-289 | PDB |
| 1NVQ | X-ray | 200 A | A | 1-289 | PDB |
| 1NVR | X-ray | 180 A | A | 1-289 | PDB |
| 1NVS | X-ray | 180 A | A | 1-289 | PDB |
| 1ZLT | X-ray | 174 A | A | 1-289 | PDB |
| 1ZYS | X-ray | 170 A | A | 1-273 | PDB |
| 2AYP | X-ray | 290 A | A | 1-269 | PDB |
| 2BR1 | X-ray | 200 A | A | 1-289 | PDB |
| 2BRB | X-ray | 210 A | A | 1-289 | PDB |
| 2BRG | X-ray | 210 A | A | 1-289 | PDB |
| 2BRH | X-ray | 210 A | A | 1-289 | PDB |
| 2BRM | X-ray | 220 A | A | 1-289 | PDB |
| 2BRN | X-ray | 280 A | A | 1-289 | PDB |
| 2BRO | X-ray | 220 A | A | 1-289 | PDB |
| 2C3J | X-ray | 210 A | A | 1-289 | PDB |
| 2C3K | X-ray | 260 A | A | 1-289 | PDB |
| 2C3L | X-ray | 235 A | A | 1-289 | PDB |
| 2CGU | X-ray | 250 A | A | 1-289 | PDB |
| 2CGV | X-ray | 260 A | A | 1-289 | PDB |
| 2CGW | X-ray | 220 A | A | 1-289 | PDB |
| 2CGX | X-ray | 220 A | A | 1-289 | PDB |
| 2E9N | X-ray | 250 A | A | 2-270 | PDB |
| 2E9O | X-ray | 210 A | A | 2-270 | PDB |
| 2E9P | X-ray | 260 A | A | 2-270 | PDB |
| 2E9U | X-ray | 200 A | A | 2-270 | PDB |
| 2E9V | X-ray | 200 A | A/B | 2-269 | PDB |
| 2GDO | X-ray | 300 A | A | 1-289 | PDB |
| 2GHG | X-ray | 350 A | A | 2-270 | PDB |
| 2HOG | X-ray | 190 A | A | 2-307 | PDB |
| 2HXL | X-ray | 180 A | A | 2-307 | PDB |
| 2HXQ | X-ray | 200 A | A | 2-307 | PDB |
| 2HY0 | X-ray | 170 A | A | 2-307 | PDB |
| 2QHM | X-ray | 200 A | A | 1-307 | PDB |
| 2QHN | X-ray | 170 A | A | 1-307 | PDB |
| 2R0U | X-ray | 190 A | A | 1-307 | PDB |
| 2WMQ | X-ray | 248 A | A | 1-289 | PDB |
| 2WMR | X-ray | 243 A | A | 1-289 | PDB |
| 2WMS | X-ray | 270 A | A | 1-289 | PDB |
| 2WMT | X-ray | 255 A | A | 1-289 | PDB |
| 2WMU | X-ray | 260 A | A | 1-289 | PDB |
| 2WMV | X-ray | 201 A | A | 1-289 | PDB |
| 2WMW | X-ray | 243 A | A | 1-289 | PDB |
| 2WMX | X-ray | 245 A | A | 1-289 | PDB |
| 2X8D | X-ray | 190 A | A | 1-289 | PDB |
| 2X8E | X-ray | 250 A | A | 1-276 | PDB |
| 2X8I | X-ray | 192 A | A | 1-289 | PDB |
| 2XEY | X-ray | 270 A | A | 1-289 | PDB |
| 2XEZ | X-ray | 225 A | A | 1-289 | PDB |
| 2XF0 | X-ray | 240 A | A | 1-289 | PDB |
| 2YDI | X-ray | 160 A | A | 1-289 | PDB |
| 2YDJ | X-ray | 185 A | A/B | 1-276 | PDB |
| 2YDK | X-ray | 190 A | A | 1-276 | PDB |
| 2YER | X-ray | 183 A | A | 1-276 | PDB |
| 2YEX | X-ray | 130 A | A | 1-276 | PDB |
| 2YM3 | X-ray | 201 A | A | 1-289 | PDB |
| 2YM4 | X-ray | 235 A | A | 1-289 | PDB |
| 2YM5 | X-ray | 203 A | A | 1-289 | PDB |
| 2YM6 | X-ray | 201 A | A | 1-289 | PDB |
| 2YM7 | X-ray | 181 A | A | 1-289 | PDB |
| 2YM8 | X-ray | 207 A | A | 1-289 | PDB |
| 2YWP | X-ray | 290 A | A | 2-270 | PDB |
| 3F9N | X-ray | 190 A | A | 2-307 | PDB |
| 3JVR | X-ray | 176 A | A | 2-272 | PDB |
| 3JVS | X-ray | 190 A | A | 2-272 | PDB |
| 3NLB | X-ray | 190 A | A | 1-289 | PDB |
| 3OT3 | X-ray | 144 A | A | 2-274 | PDB |
| 3OT8 | X-ray | 165 A | A | 2-274 | PDB |
| 3PA3 | X-ray | 140 A | A | 2-274 | PDB |
| 3PA4 | X-ray | 159 A | A | 2-274 | PDB |
| 3PA5 | X-ray | 170 A | A | 2-274 | PDB |
| 3TKH | X-ray | 179 A | A | 1-307 | PDB |
| 3TKI | X-ray | 160 A | A | 1-307 | PDB |
| 3U9N | X-ray | 185 A | A | 2-274 | PDB |
| 4FSM | X-ray | 230 A | A | 2-280 | PDB |
| 4FSN | X-ray | 210 A | A | 4-280 | PDB |
| 4FSQ | X-ray | 240 A | A | 2-280 | PDB |
| 4FSR | X-ray | 250 A | A | 2-280 | PDB |
| 4FST | X-ray | 190 A | A | 2-270 | PDB |
| 4FSU | X-ray | 210 A | A | 2-280 | PDB |
| 4FSW | X-ray | 230 A | A | 2-280 | PDB |
| 4FSY | X-ray | 230 A | A | 2-280 | PDB |
| 4FSZ | X-ray | 230 A | A | 2-280 | PDB |
| 4FT0 | X-ray | 230 A | A | 2-280 | PDB |
| 4FT3 | X-ray | 250 A | A | 2-280 | PDB |
| 4FT5 | X-ray | 240 A | A | 2-280 | PDB |
| 4FT7 | X-ray | 220 A | A | 2-280 | PDB |
| 4FT9 | X-ray | 220 A | A | 2-280 | PDB |
| 4FTA | X-ray | 240 A | A | 2-280 | PDB |
| 4FTC | X-ray | 200 A | A | 2-280 | PDB |
| 4FTI | X-ray | 220 A | A | 2-280 | PDB |
| 4FTJ | X-ray | 220 A | A | 2-280 | PDB |
| 4FTK | X-ray | 230 A | A | 2-280 | PDB |
| 4FTL | X-ray | 250 A | A | 2-280 | PDB |
| 4FTM | X-ray | 190 A | A | 2-280 | PDB |
| 4FTN | X-ray | 202 A | A | 2-280 | PDB |
| 4FTO | X-ray | 210 A | A | 2-280 | PDB |
| 4FTQ | X-ray | 200 A | A | 2-280 | PDB |
| 4FTR | X-ray | 225 A | A | 2-280 | PDB |
| 4FTT | X-ray | 230 A | A | 2-280 | PDB |
| 4FTU | X-ray | 210 A | A | 2-280 | PDB |
| 4GH2 | X-ray | 203 A | A | 2-280 | PDB |
| 4HYH | X-ray | 170 A | A | 1-289 | PDB |
| 4HYI | X-ray | 140 A | A | 1-289 | PDB |
| 4JIK | X-ray | 190 A | A | 2-274 | PDB |
| 4QYE | X-ray | 205 A | A | 1-289 | PDB |
| 4QYF | X-ray | 215 A | A | 1-289 | PDB |
| 4QYG | X-ray | 175 A | A/B | 1-289 | PDB |
| 4QYH | X-ray | 190 A | A/B | 1-289 | PDB |
| 4RVK | X-ray | 185 A | A | 1-289 | PDB |
| 4RVL | X-ray | 185 A | A | 1-289 | PDB |
| 4RVM | X-ray | 186 A | A | 1-289 | PDB |
| 5DLS | X-ray | 215 A | A | 1-289 | PDB |
| 5F4N | X-ray | 191 A | A | 1-273 | PDB |
| 5OOP | X-ray | 170 A | A | 1-289 | PDB |
| 5OOR | X-ray | 190 A | A | 1-289 | PDB |
| 5OOT | X-ray | 210 A | A | 1-289 | PDB |
| 5OP2 | X-ray | 190 A | A | 1-289 | PDB |
| 5OP4 | X-ray | 200 A | A | 1-289 | PDB |
| 5OP5 | X-ray | 190 A | A | 1-289 | PDB |
| 5OP7 | X-ray | 180 A | A | 1-289 | PDB |
| 5OPB | X-ray | 155 A | A | 1-289 | PDB |
| 5OPR | X-ray | 195 A | A | 1-289 | PDB |
| 5OPS | X-ray | 200 A | A | 1-289 | PDB |
| 5OPU | X-ray | 155 A | A | 1-289 | PDB |
| 5OPV | X-ray | 190 A | A | 1-289 | PDB |
| 5OQ5 | X-ray | 140 A | A | 1-289 | PDB |
| 5OQ6 | X-ray | 195 A | A | 1-289 | PDB |
| 5OQ7 | X-ray | 210 A | A/B | 1-289 | PDB |
| 5OQ8 | X-ray | 200 A | A | 1-289 | PDB |
| 5WI2 | X-ray | 250 A | A/B | 377-476 | PDB |
| 6FC8 | X-ray | 161 A | A | 1-276 | PDB |
| 6FCF | X-ray | 185 A | A | 1-276 | PDB |
| 6FCK | X-ray | 190 A | A | 1-276 | PDB |
| 7AKM | X-ray | 193 A | A/B | 2-287 | PDB |
| 7AKO | X-ray | 180 A | A/B | 2-289 | PDB |
| 7BJD | X-ray | 200 A | A | 1-289 | PDB |
| 7BJE | X-ray | 180 A | A | 1-289 | PDB |
| 7BJH | X-ray | 180 A | A | 1-289 | PDB |
| 7BJJ | X-ray | 180 A | A | 1-289 | PDB |
| 7BJM | X-ray | 230 A | A | 1-289 | PDB |
| 7BJO | X-ray | 230 A | A | 1-289 | PDB |
| 7BJR | X-ray | 190 A | A | 1-289 | PDB |
| 7BJX | X-ray | 240 A | A/B | 1-289 | PDB |
| 7BK1 | X-ray | 200 A | A | 1-289 | PDB |
| 7BK2 | X-ray | 200 A | A | 1-289 | PDB |
| 7BK3 | X-ray | 200 A | A | 1-289 | PDB |
| 7BKN | X-ray | 274 A | A | 1-289 | PDB |
| 7BKO | X-ray | 230 A | A | 1-289 | PDB |
| 7MCK | X-ray | 165 A | A | 1-289 | PDB |
| 7SUF | X-ray | 148 A | A | 1-289 | PDB |
| 7SUG | X-ray | 148 A | A | 1-289 | PDB |
| 7SUH | X-ray | 246 A | A | 1-289 | PDB |
| 7SUI | X-ray | 212 A | A | 1-289 | PDB |
| 7SUJ | X-ray | 230 A | A/B | 1-289 | PDB |
| 8E80 | X-ray | 149 A | A | 1-289 | PDB |
| 8E81 | X-ray | 162 A | A | 1-289 | PDB |
| 8SIV | X-ray | 176 A | A | 1-289 | PDB |
| 8SIW | X-ray | 188 A | A/B | 1-289 | PDB |
| 8SIX | X-ray | 155 A | A | 1-289 | PDB |
| AF-O14757-F1 | Predicted | AlphaFoldDB |
276 variants for O14757
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA6349430 rs369248914 RCV000708693 |
160 | R>C | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA351245 rs756054805 RCV000210167 |
379 | R>* | Hereditary cancer-predisposing syndrome [ClinVar] | Yes |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
CA6349285 rs757321404 |
2 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA6349286 rs201459198 |
4 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs200520758 CA6349287 |
12 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383189905 rs1395953036 |
18 | G>D | No |
ClinGen TOPMed |
|
|
rs1350704599 CA383189936 |
20 | Y>C | No |
ClinGen gnomAD |
|
|
rs751896993 CA6349290 |
21 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA6349308 rs766554164 |
25 | L>I | No |
ClinGen ExAC gnomAD |
|
|
rs1565361701 CA383190120 |
26 | A>V | No |
ClinGen Ensembl |
|
|
CA6349310 rs755282936 |
28 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA383190150 rs1555067411 RCV000586016 |
29 | R>G | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA6349311 rs781671034 |
30 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA383190178 rs1191531722 |
31 | T>N | No |
ClinGen gnomAD |
|
|
rs1269445439 CA383190213 |
34 | A>T | No |
ClinGen gnomAD |
|
|
rs1380056031 CA383190227 |
35 | V>L | No |
ClinGen TOPMed |
|
|
rs753180922 CA6349312 |
36 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs753180922 CA6349313 |
36 | A>T | No |
ClinGen ExAC gnomAD |
|
|
CA6349314 rs367942205 |
39 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs778668643 CA6349317 |
41 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383190301 rs1458791146 |
42 | M>V | No |
ClinGen gnomAD |
|
|
rs771939046 CA6349319 |
44 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs537858020 CA6349320 |
44 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA6349321 rs760734978 |
45 | A>D | No |
ClinGen ExAC gnomAD |
|
|
rs1565361807 CA383190322 |
45 | A>S | No |
ClinGen Ensembl |
|
|
rs768467449 CA6349322 |
46 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1367390698 CA383190345 |
48 | C>* | No |
ClinGen gnomAD |
|
|
CA383190365 rs1565361841 |
51 | N>S | No |
ClinGen Ensembl |
|
|
rs779362881 CA230484454 |
55 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762977718 CA6349324 |
55 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1266930698 CA383190407 |
57 | C>G | No |
ClinGen gnomAD |
|
|
CA383190442 rs1248538225 |
61 | M>I | No |
ClinGen gnomAD |
|
|
rs751719325 CA6349331 |
64 | H>N | No |
ClinGen ExAC gnomAD |
|
|
CA383190471 rs554220572 |
65 | E>D | No |
ClinGen 1000Genomes ExAC |
|
|
rs554220572 CA6349333 |
65 | E>D | No |
ClinGen 1000Genomes ExAC |
|
|
CA6349335 rs767992502 |
67 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs767992502 CA6349336 |
67 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs375481689 CA6349339 |
69 | K>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1386843282 CA383190495 |
69 | K>N | No |
ClinGen gnomAD |
|
|
rs1356561612 CA383190499 |
70 | F>V | No |
ClinGen TOPMed |
|
|
CA6349340 rs141665265 |
71 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA383190509 rs141665265 |
71 | Y>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA383190519 rs1224598742 |
73 | H>D | No |
ClinGen TOPMed |
|
|
CA383190535 rs1565362006 |
75 | R>K | No |
ClinGen Ensembl |
|
|
rs758124077 CA6349343 |
77 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6349345 rs746760826 |
79 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs768667116 CA6349346 |
80 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1227533180 CA383190593 |
83 | F>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs1207424665 CA383190602 |
85 | E>K | No |
ClinGen gnomAD |
|
|
CA6349352 rs767610516 |
87 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA6349353 rs775964760 |
92 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs764684778 CA6349355 |
96 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs754338984 CA6349356 |
97 | E>* | No |
ClinGen ExAC gnomAD |
|
|
CA383190728 rs1426460534 |
101 | G>C | No |
ClinGen TOPMed gnomAD |
|
|
CA383190726 rs1426460534 |
101 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs764300223 CA6349374 |
106 | D>H | No |
ClinGen ExAC gnomAD |
|
|
rs764300223 CA6349375 |
106 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs762470361 CA6349376 |
107 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs563722813 CA6349377 |
111 | F>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1383121355 CA383190804 |
112 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA383190824 rs1197536561 |
115 | M>V | No |
ClinGen TOPMed |
|
|
CA230484942 rs1048249612 |
117 | G>R | No |
ClinGen Ensembl |
|
|
rs749996823 CA6349378 |
118 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs1207337706 CA383190893 |
124 | I>V | No |
ClinGen gnomAD |
|
|
rs112275638 CA230485003 |
126 | I>V | No |
ClinGen Ensembl |
|
|
rs867675787 CA230485006 |
129 | R>G | No |
ClinGen Ensembl |
|
|
CA6349400 rs759158547 |
137 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA6349401 rs752519129 |
137 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6349402 rs371765740 |
139 | D>Y | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1391121414 CA383191009 |
141 | R>K | No |
ClinGen gnomAD |
|
|
COSM541508 CA383191012 rs777701208 |
141 | R>S | lung [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs1279298605 CA383191271 |
144 | L>R | No |
ClinGen TOPMed |
|
|
CA6349420 rs775066373 |
145 | K>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs760469704 CA6349421 |
145 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6349422 rs763944664 |
149 | F>V | No |
ClinGen ExAC gnomAD |
|
|
CA6349424 rs757193229 |
153 | T>K | No |
ClinGen ExAC gnomAD |
|
|
CA6349423 rs753657060 |
153 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs3731410 CA6349427 VAR_021123 |
156 | R>Q | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
COSM3415648 rs140276570 CA6349426 |
156 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA383191367 rs1591395808 |
157 | Y>D | No |
ClinGen Ensembl |
|
|
CA6349429 rs748445411 |
157 | Y>F | No |
ClinGen ExAC gnomAD |
|
|
CA383191381 rs1161248510 |
159 | N>D | No |
ClinGen gnomAD |
|
|
CA230486291 rs907672030 |
159 | N>I | No |
ClinGen gnomAD |
|
|
rs907672030 CA230486289 |
159 | N>S | No |
ClinGen gnomAD |
|
|
COSM35410 CA6349431 rs778046191 |
160 | R>H | large_intestine central_nervous_system endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA6349432 rs749631457 |
162 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs1427360758 CA383191415 |
162 | R>H | No |
ClinGen gnomAD |
|
|
rs771372037 CA6349433 |
164 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA6349434 rs373714183 COSM924552 |
166 | K>N | endometrium [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1299890343 CA383191511 |
167 | M>I | No |
ClinGen gnomAD |
|
|
rs1393469843 CA383191500 |
167 | M>L | No |
ClinGen gnomAD |
|
|
rs1393469843 CA383191496 |
167 | M>L | No |
ClinGen gnomAD |
|
|
CA6349435 rs745384189 |
168 | C>G | No |
ClinGen ExAC gnomAD |
|
|
CA383191566 rs1234061120 |
171 | L>S | No |
ClinGen gnomAD |
|
|
CA383191579 rs1340501726 |
172 | P>S | No |
ClinGen gnomAD |
|
|
CA383191625 rs1158434447 |
174 | V>A | No |
ClinGen TOPMed |
|
|
CA383191616 rs1378781957 |
174 | V>L | No |
ClinGen TOPMed |
|
|
CA383191636 rs1340329656 |
175 | A>S | No |
ClinGen gnomAD |
|
|
rs75583143 CA230486314 |
175 | A>V | No |
ClinGen Ensembl |
|
|
CA230486317 rs1037595364 |
176 | P>L | No |
ClinGen Ensembl |
|
|
rs755263451 CA6349436 |
180 | K>L | No |
ClinGen ExAC |
|
|
CA6349439 rs771504275 |
180 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1591395964 CA383191725 |
181 | R>G | No |
ClinGen Ensembl |
|
|
CA230486327 rs995839011 |
183 | E>G | No |
ClinGen gnomAD |
|
|
CA6349440 rs760379838 |
185 | H>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1396311668 CA383191868 |
189 | V>A | No |
ClinGen TOPMed gnomAD |
|
|
rs776333699 CA6349442 |
191 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA383191947 rs1167618904 |
194 | C>R | No |
ClinGen TOPMed gnomAD |
|
|
CA383192001 rs1463911220 |
196 | I>T | No |
ClinGen gnomAD |
|
|
CA6349445 rs765113622 |
197 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs1363161744 CA383192088 |
200 | A>G | No |
ClinGen gnomAD |
|
|
CA6349448 rs771316687 |
201 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs376741119 CA6349450 |
203 | A>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA6349451 rs778014033 |
205 | E>* | No |
ClinGen ExAC gnomAD |
|
|
CA6349476 rs779394398 |
210 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs547707890 CA6349477 |
211 | P>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA230487164 rs901928346 |
213 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs377704459 CA6349478 |
216 | Q>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1445875437 CA383193276 |
220 | D>E | No |
ClinGen gnomAD |
|
|
CA6349479 rs780501566 |
220 | D>Y | No |
ClinGen ExAC gnomAD |
|
|
CA383193284 rs1402782439 |
221 | W>* | No |
ClinGen gnomAD |
|
|
rs35817404 CA383193299 |
223 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs35817404 CA6349482 VAR_040407 |
223 | E>V | No |
ClinGen UniProt ExAC TOPMed dbSNP gnomAD |
|
|
rs1266487380 CA383193307 |
224 | K>I | No |
ClinGen TOPMed |
|
|
rs1042455164 CA230487180 |
225 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1327022943 CA383193329 |
227 | Y>* | No |
ClinGen gnomAD |
|
|
CA6349486 rs769305503 |
228 | L>F | No |
ClinGen ExAC gnomAD |
|
|
CA6349487 rs773178839 |
229 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA6349488 rs762794644 |
230 | P>A | No |
ClinGen ExAC gnomAD |
|
|
rs1011779266 CA383193379 |
234 | I>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1255118863 CA383193380 COSM924554 |
235 | D>N | endometrium [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
rs1190534193 CA383193400 |
238 | P>S | No |
ClinGen gnomAD |
|
|
CA230487844 rs975304527 |
243 | H>N | No |
ClinGen TOPMed |
|
|
CA383193441 rs975304527 |
243 | H>Y | No |
ClinGen TOPMed |
|
|
rs777516943 CA6349505 |
245 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA383193467 rs770782396 |
246 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs201395400 CA6349508 |
251 | S>P | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA383193503 rs1158338803 |
252 | A>S | No |
ClinGen TOPMed |
|
|
CA383193506 rs1434154060 |
252 | A>V | No |
ClinGen gnomAD |
|
|
CA383193517 rs1302103336 |
254 | I>T | No |
ClinGen gnomAD |
|
|
rs11553952 CA230487861 |
254 | I>V | No |
ClinGen Ensembl |
|
|
rs745876688 CA6349509 |
255 | T>I | No |
ClinGen ExAC gnomAD |
|
|
rs771882988 CA6349510 |
257 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA383193539 rs201385540 |
258 | D>H | No |
ClinGen gnomAD |
|
|
rs201385540 CA230487870 |
258 | D>N | No |
ClinGen gnomAD |
|
|
rs1275492676 CA383193567 |
261 | K>N | No |
ClinGen gnomAD |
|
|
rs1215535083 CA383193564 |
261 | K>R | No |
ClinGen gnomAD |
|
|
CA383193570 rs1591400664 |
262 | D>H | No |
ClinGen Ensembl |
|
|
rs1450424448 CA383193578 |
263 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1470286568 CA383193588 |
264 | W>* | No |
ClinGen gnomAD |
|
|
CA230487877 rs922245598 |
264 | W>R | No |
ClinGen gnomAD |
|
|
CA383193606 rs1254148750 |
266 | N>K | No |
ClinGen gnomAD |
|
|
CA383193611 rs1469921398 |
267 | K>R | No |
ClinGen gnomAD |
|
|
CA230487882 rs933668612 |
269 | L>H | No |
ClinGen TOPMed gnomAD |
|
|
CA383193638 rs1389785450 |
271 | K>R | No |
ClinGen gnomAD |
|
|
rs1475833328 CA383193644 |
272 | G>W | No |
ClinGen gnomAD |
|
|
CA383187637 rs1383668839 |
273 | A>E | No |
ClinGen gnomAD |
|
|
CA6349530 rs778901513 |
273 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1382274821 CA383187642 |
274 | K>* | No |
ClinGen gnomAD |
|
|
rs1382274821 CA383187641 |
274 | K>E | No |
ClinGen gnomAD |
|
|
rs1308818250 CA383187644 |
274 | K>R | No |
ClinGen gnomAD |
|
|
rs745620670 CA6349531 |
275 | R>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383187652 rs1351213254 |
275 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
CA6349532 rs772079899 |
277 | R>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383187660 rs772079899 |
277 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775245290 CA6349533 |
277 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs748276359 CA6349534 |
282 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA6349535 rs748276359 |
282 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA383187691 rs1227056031 |
283 | V>M | No |
ClinGen gnomAD |
|
|
CA383187707 rs1260622411 |
285 | E>G | No |
ClinGen gnomAD |
|
|
CA383187722 rs1314732435 |
287 | P>L | No |
ClinGen TOPMed |
|
|
rs1355494579 CA383187719 |
287 | P>S | No |
ClinGen gnomAD |
|
|
rs763207260 CA383187727 |
288 | S>I | No |
ClinGen ExAC gnomAD |
|
|
rs763207260 CA6349537 |
288 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1485430319 CA383187730 |
289 | G>R | No |
ClinGen gnomAD |
|
|
rs147323405 CA230472755 |
293 | H>Y | No |
ClinGen ESP |
|
|
CA383187774 rs1186977956 |
295 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
CA383187772 rs1186977956 |
295 | Q>E | No |
ClinGen TOPMed gnomAD |
|
|
CA230472760 rs1003827905 |
297 | N>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1179637125 CA383187820 |
301 | S>F | No |
ClinGen gnomAD |
|
|
CA230472763 rs1025553804 |
301 | S>P | No |
ClinGen Ensembl |
|
|
rs1363144435 CA383187826 |
302 | P>L | No |
ClinGen gnomAD |
|
|
CA6349539 rs182542971 |
304 | N>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1159724450 CA383187846 |
305 | S>R | No |
ClinGen gnomAD |
|
|
CA383187850 rs1399624091 |
306 | A>S | No |
ClinGen gnomAD |
|
|
rs1433658665 CA383187853 |
306 | A>V | No |
ClinGen gnomAD |
|
|
rs1437919116 CA383187859 |
307 | S>F | No |
ClinGen gnomAD |
|
|
CA230472859 VAR_040408 rs34097480 |
312 | V>M | No |
ClinGen UniProt Ensembl dbSNP |
|
|
CA383187916 rs1322109311 |
313 | K>N | No |
ClinGen gnomAD |
|
|
rs1370402981 CA383187919 |
314 | Y>D | No |
ClinGen gnomAD |
|
|
rs1398654305 CA383187932 |
316 | S>G | No |
ClinGen gnomAD |
|
|
rs1235097713 CA383187934 |
316 | S>N | No |
ClinGen gnomAD |
|
|
CA383187972 rs1298017021 |
321 | P>L | No |
ClinGen TOPMed |
|
|
rs1340219994 CA383187967 |
321 | P>T | No |
ClinGen gnomAD |
|
|
rs775909758 CA6349562 |
322 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6349563 COSM1352378 rs761313560 |
322 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
CA383187976 rs761313560 |
322 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383187980 rs764666835 |
323 | T>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764666835 CA6349564 |
323 | T>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs753432559 CA6349565 |
324 | G>S | No |
ClinGen ExAC gnomAD |
|
|
CA230472882 rs1026455651 |
328 | W>C | No |
ClinGen TOPMed |
|
|
rs764958538 CA6349569 |
333 | S>* | No |
ClinGen ExAC gnomAD |
|
|
rs764958538 CA383188046 |
333 | S>L | No |
ClinGen ExAC gnomAD |
|
|
rs1426308049 CA383188042 |
333 | S>T | No |
ClinGen gnomAD |
|
|
CA6349570 rs749980029 |
334 | Y>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA230472890 rs781018582 |
334 | Y>C | No |
ClinGen gnomAD |
|
|
CA383188049 rs781018582 |
334 | Y>F | No |
ClinGen gnomAD |
|
|
CA230472888 rs139936669 |
334 | Y>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1441734268 CA383188055 |
335 | I>T | No |
ClinGen gnomAD |
|
|
rs758069861 CA6349571 |
335 | I>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs539323916 CA6349575 |
341 | G>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA6349576 rs749381202 |
342 | I>L | No |
ClinGen ExAC gnomAD |
|
|
rs1591409094 CA383188110 |
343 | S>T | No |
ClinGen Ensembl |
|
|
rs199535573 CA501216 |
346 | Q>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs374866866 CA6349579 |
348 | T>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs761070355 CA6349581 |
350 | P>H | No |
ClinGen ExAC gnomAD |
|
|
CA6349580 rs775766777 |
350 | P>S | No |
ClinGen ExAC gnomAD |
|
|
COSM107213 rs149667923 CA230472919 |
352 | H>Y | skin [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
rs777209903 CA6349583 |
353 | M>I | No |
ClinGen ExAC |
|
|
rs761418252 CA6349584 |
356 | N>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1163144259 CA383188206 |
357 | S>T | No |
ClinGen gnomAD |
|
|
rs1207019231 CA383188210 |
358 | Q>E | No |
ClinGen TOPMed |
|
|
rs1462376220 CA383188241 |
362 | T>S | No |
ClinGen gnomAD |
|
|
rs764708405 CA6349585 |
363 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA6349586 rs764708405 |
363 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs542436508 CA230472939 |
366 | S>L | No |
ClinGen Ensembl |
|
|
CA6349607 rs773937190 |
368 | N>K | No |
ClinGen ExAC gnomAD |
|
|
rs867390301 CA230473027 |
368 | N>S | No |
ClinGen Ensembl |
|
|
rs1318383609 CA383188291 |
369 | P>S | No |
ClinGen gnomAD |
|
|
rs1279132447 CA383188298 |
370 | W>* | No |
ClinGen gnomAD |
|
|
CA230473035 rs77764256 |
370 | W>C | No |
ClinGen Ensembl |
|
|
CA383188297 rs1443980454 |
370 | W>G | No |
ClinGen TOPMed |
|
|
rs1486238508 CA383188305 |
371 | Q>* | No |
ClinGen gnomAD |
|
|
rs767367809 CA6349609 |
372 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383188332 rs1183852401 |
375 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
CA6349610 rs752458006 |
377 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA6349612 rs750498287 |
385 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs1395913974 CA383188513 |
386 | A>T | No |
ClinGen gnomAD |
|
|
CA6349613 rs758434651 |
389 | S>F | No |
ClinGen ExAC |
|
|
rs372751798 CA230473054 |
390 | Y>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1257217942 CA383188605 |
390 | Y>F | No |
ClinGen gnomAD |
|
|
rs1435108441 CA383188602 |
390 | Y>H | No |
ClinGen gnomAD |
|
|
CA6349615 rs747288732 |
395 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA6349618 rs748478311 |
397 | C>F | No |
ClinGen ExAC gnomAD |
|
|
rs1318978818 CA383188881 |
401 | G>A | No |
ClinGen TOPMed |
|
|
CA230473068 rs115991933 |
403 | Q>R | No |
ClinGen 1000Genomes gnomAD |
|
|
rs748763845 CA6349621 |
404 | W>C | No |
ClinGen ExAC gnomAD |
|
|
rs145794689 CA6349623 |
406 | K>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1403145263 CA383189030 |
408 | C>S | No |
ClinGen TOPMed |
|
|
CA6349624 rs759227173 |
409 | M>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA383189104 rs1455708220 |
411 | Q>P | No |
ClinGen gnomAD |
|
|
CA230474784 rs753915531 |
412 | V>I | No |
ClinGen Ensembl |
|
|
CA6349639 rs749874631 |
413 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs148568641 CA6349640 |
414 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs761805263 CA230474787 |
415 | S>T | No |
ClinGen Ensembl |
|
|
CA6349641 rs778444034 |
417 | T>N | No |
ClinGen ExAC gnomAD |
|
|
rs370268372 CA6349643 |
421 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA383191095 rs775056015 |
422 | N>K | No |
ClinGen ExAC gnomAD |
|
|
CA230474793 rs914262719 |
434 | D>V | No |
ClinGen Ensembl |
|
|
CA6349646 rs768550804 |
439 | V>A | No |
ClinGen ExAC gnomAD |
|
|
CA230474797 rs968420616 |
442 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
rs751724890 CA6349650 |
445 | K>E | No |
ClinGen ExAC gnomAD |
|
|
rs1055671859 CA230475067 |
457 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1439749537 CA383191818 |
462 | L>V | No |
ClinGen gnomAD |
|
|
CA6349673 rs775403114 |
463 | I>M | No |
ClinGen ExAC gnomAD |
|
|
rs761007676 CA6349674 |
464 | D>A | No |
ClinGen ExAC gnomAD |
|
|
CA6349676 rs529219511 |
465 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA6349675 rs764175263 |
465 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA6349677 rs146590504 |
466 | V>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs750985875 CA6349679 |
469 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
rs506504 CA230475078 |
471 | I>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs506504 CA230475076 |
471 | I>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs506504 CA6349680 RCV000589283 VAR_024571 |
471 | I>V | No |
ClinGen ClinVar UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1242394353 CA383192090 |
473 | L>P | No |
ClinGen gnomAD |
|
|
rs779651907 CA6349681 |
476 | T>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1202783491 CA383192159 |
476 | T>I | No |
ClinGen TOPMed |
No associated diseases with O14757
6 regional properties for O14757
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 493 - 770 | IPR000719 |
| domain | S-locus glycoprotein domain | 209 - 319 | IPR000858 |
| domain | Bulb-type lectin domain | 23 - 178 | IPR001480 |
| domain | PAN/Apple domain | 341 - 422 | IPR003609 |
| active_site | Serine/threonine-protein kinase, active site | 615 - 627 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 499 - 521 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR24346 | MAP/MICROTUBULE AFFINITY-REGULATING KINASE |
| PANTHER Subfamily | PTHR24346:SF73 | SERINE_THREONINE-PROTEIN KINASE CHK1 |
| PANTHER Protein Class |
non-receptor serine/threonine protein kinase
protein modifying enzyme |
|
| PANTHER Pathway Category | No pathway information available | |
11 GO annotations of cellular component
| Name | Definition |
|---|---|
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| condensed nuclear chromosome | A highly compacted molecule of DNA and associated proteins resulting in a cytologically distinct nuclear chromosome. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular space | That part of a multicellular organism outside the cells proper, usually taken to be outside the plasma membranes, and occupied by fluid. |
| intracellular membrane-bounded organelle | Organized structure of distinctive morphology and function, bounded by a single or double lipid bilayer membrane and occurring within the cell. Includes the nucleus, mitochondria, plastids, vacuoles, and vesicles. Excludes the plasma membrane. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| protein-containing complex | A stable assembly of two or more macromolecules, i.e. proteins, nucleic acids, carbohydrates or lipids, in which at least one component is a protein and the constituent parts function together. |
| replication fork | The Y-shaped region of a replicating DNA molecule, resulting from the separation of the DNA strands and in which the synthesis of new strands takes place. Also includes associated protein complexes. |
7 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| histone kinase activity (H3-T11 specific) | Catalysis of the transfer of a phosphate group to the threonine-11 residue of the N-terminal tail of histone H3. |
| protein domain specific binding | Binding to a specific domain of a protein. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
29 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| apoptotic process involved in development | Any apoptotic process that is involved in anatomical structure development. |
| cellular response to caffeine | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a caffeine stimulus. Caffeine is an alkaloid found in numerous plant species, where it acts as a natural pesticide that paralyzes and kills certain insects feeding upon them. |
| cellular response to DNA damage stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to its DNA from environmental insults or errors during metabolism. |
| cellular response to mechanical stimulus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a mechanical stimulus. |
| DNA damage checkpoint signaling | A signal transduction process that contributes to a DNA damage checkpoint. |
| DNA damage induced protein phosphorylation | The widespread phosphorylation of various molecules, triggering many downstream processes, that occurs in response to the detection of DNA damage. |
| DNA repair | The process of restoring DNA after damage. Genomes are subject to damage by chemical and physical agents in the environment (e.g. UV and ionizing radiations, chemical mutagens, fungal and bacterial toxins, etc.) and by free radicals or alkylating agents endogenously generated in metabolism. DNA is also damaged because of errors during its replication. A variety of different DNA repair pathways have been reported that include direct reversal, base excision repair, nucleotide excision repair, photoreactivation, bypass, double-strand break repair pathway, and mismatch repair pathway. |
| DNA replication | The cellular metabolic process in which a cell duplicates one or more molecules of DNA. DNA replication begins when specific sequences, known as origins of replication, are recognized and bound by initiation proteins, and ends when the original DNA molecule has been completely duplicated and the copies topologically separated. The unit of replication usually corresponds to the genome of the cell, an organelle, or a virus. The template for replication can either be an existing DNA molecule or RNA. |
| epigenetic maintenance of chromatin in transcription-competent conformation | An epigenetic process that capacitates gene expression by remodelling of chromatin by either modifying the chromatin fiber, the nucleosomal histones, or the DNA. |
| G2/M transition of mitotic cell cycle | The mitotic cell cycle transition by which a cell in G2 commits to M phase. The process begins when the kinase activity of M cyclin/CDK complex reaches a threshold high enough for the cell cycle to proceed. This is accomplished by activating a positive feedback loop that results in the accumulation of unphosphorylated and active M cyclin/CDK complex. |
| inner cell mass cell proliferation | The proliferation of cells in the inner cell mass. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| mitotic G2 DNA damage checkpoint signaling | A mitotic cell cycle checkpoint that detects and negatively regulates progression through the G2/M transition of the cell cycle in response to DNA damage. |
| mitotic G2/M transition checkpoint | A cell cycle checkpoint that detects and negatively regulates progression from G2 to M phase as part of a mitotic cell cycle. |
| negative regulation of DNA biosynthetic process | Any process that stops, prevents or reduces the frequency, rate or extent of DNA biosynthetic process. |
| negative regulation of G0 to G1 transition | A cell cycle process that stops, prevents, or reduces the rate or extent of the transition from the G0 quiescent state to the G1 phase. |
| negative regulation of mitotic nuclear division | Any process that stops, prevents or reduces the rate or extent of mitosis. Mitosis is the division of the eukaryotic cell nucleus to produce two daughter nuclei that, usually, contain the identical chromosome complement to their mother. |
| nucleus organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the nucleus. |
| peptidyl-threonine phosphorylation | The phosphorylation of peptidyl-threonine to form peptidyl-O-phospho-L-threonine. |
| positive regulation of cell cycle | Any process that activates or increases the rate or extent of progression through the cell cycle. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of cell population proliferation | Any process that modulates the frequency, rate or extent of cell proliferation. |
| regulation of double-strand break repair via homologous recombination | Any process that modulates the frequency, rate or extent of the error-free repair of a double-strand break in DNA in which the broken DNA molecule is repaired using homologous sequences. |
| regulation of histone H3-K9 acetylation | Any process that modulates the frequency, rate or extent of histone H3-K9 acetylation. |
| regulation of mitotic centrosome separation | Any process that modulates the frequency, rate or extent of the separation of duplicated centrosome components at the beginning of mitosis. |
| regulation of signal transduction by p53 class mediator | Any process that modulates the frequency, rate or extent of signal transduction by p53 class mediator. |
| regulation of transcription from RNA polymerase II promoter in response to UV-induced DNA damage | Any process that modulates the frequency, rate or extent of transcription from an RNA polymerase II promoter as a result of a UV damage stimulus. |
| replicative senescence | A cell aging process associated with the dismantling of a cell as a response to telomere shortening and/or cellular aging. |
21 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28708 | PRR1 | Serine/threonine-protein kinase PRR1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P47116 | PTK2 | Serine/threonine-protein kinase PTK2/STK2 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q8AYC9 | CHEK1 | Serine/threonine-protein kinase Chk1 | Gallus gallus (Chicken) | PR |
| O61661 | grp | Serine/threonine-protein kinase grp | Drosophila melanogaster (Fruit fly) | SS |
| Q8IWQ3 | BRSK2 | Serine/threonine-protein kinase BRSK2 | Homo sapiens (Human) | PR |
| Q8TDC3 | BRSK1 | Serine/threonine-protein kinase BRSK1 | Homo sapiens (Human) | SS |
| Q14680 | MELK | Maternal embryonic leucine zipper kinase | Homo sapiens (Human) | EV |
| P57059 | SIK1 | Serine/threonine-protein kinase SIK1 | Homo sapiens (Human) | PR |
| Q9NRH2 | SNRK | SNF-related serine/threonine-protein kinase | Homo sapiens (Human) | SS |
| Q9BXA7 | TSSK1B | Testis-specific serine/threonine-protein kinase 1 | Homo sapiens (Human) | PR |
| O60285 | NUAK1 | NUAK family SNF1-like kinase 1 | Homo sapiens (Human) | PR |
| Q13131 | PRKAA1 | 5'-AMP-activated protein kinase catalytic subunit alpha-1 | Homo sapiens (Human) | EV |
| P54646 | PRKAA2 | 5'-AMP-activated protein kinase catalytic subunit alpha-2 | Homo sapiens (Human) | EV |
| O35280 | Chek1 | Serine/threonine-protein kinase Chk1 | Mus musculus (Mouse) | PR |
| Q91ZN7 | Chek1 | Serine/threonine-protein kinase Chk1 | Rattus norvegicus (Rat) | PR |
| Q8LIG4 | CIPK3 | CBL-interacting protein kinase 3 | Oryza sativa subsp japonica (Rice) | PR |
| Q5JLQ9 | CIPK30 | CBL-interacting protein kinase 30 | Oryza sativa subsp japonica (Rice) | PR |
| Q6ERS4 | CIPK16 | CBL-interacting protein kinase 16 | Oryza sativa subsp japonica (Rice) | PR |
| Q6ZLP5 | CIPK23 | CBL-interacting protein kinase 23 | Oryza sativa subsp japonica (Rice) | PR |
| Q2QY53 | CIPK32 | CBL-interacting protein kinase 32 | Oryza sativa subsp japonica (Rice) | PR |
| Q2RAX3 | CIPK33 | CBL-interacting protein kinase 33 | Oryza sativa subsp japonica (Rice) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAVPFVEDWD | LVQTLGEGAY | GEVQLAVNRV | TEEAVAVKIV | DMKRAVDCPE | NIKKEICINK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| MLNHENVVKF | YGHRREGNIQ | YLFLEYCSGG | ELFDRIEPDI | GMPEPDAQRF | FHQLMAGVVY |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LHGIGITHRD | IKPENLLLDE | RDNLKISDFG | LATVFRYNNR | ERLLNKMCGT | LPYVAPELLK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RREFHAEPVD | VWSCGIVLTA | MLAGELPWDQ | PSDSCQEYSD | WKEKKTYLNP | WKKIDSAPLA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LLHKILVENP | SARITIPDIK | KDRWYNKPLK | KGAKRPRVTS | GGVSESPSGF | SKHIQSNLDF |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SPVNSASSEE | NVKYSSSQPE | PRTGLSLWDT | SPSYIDKLVQ | GISFSQPTCP | DHMLLNSQLL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GTPGSSQNPW | QRLVKRMTRF | FTKLDADKSY | QCLKETCEKL | GYQWKKSCMN | QVTISTTDRR |
| 430 | 440 | 450 | 460 | 470 | |
| NNKLIFKVNL | LEMDDKILVD | FRLSKGDGLE | FKRHFLKIKG | KLIDIVSSQK | IWLPAT |