O14745
Gene name |
SLC9A3R1 |
Protein name |
Na(+)/H(+) exchange regulatory cofactor NHE-RF1 |
Names |
NHERF-1, Ezrin-radixin-moesin-binding phosphoprotein 50, EBP50, Regulatory cofactor of Na(+)/H(+) exchanger, Sodium-hydrogen exchanger regulatory factor 1, Solute carrier family 9 isoform A3 regulatory factor 1 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:9368 |
EC number |
|
Protein Class |
PDZ DOMAIN CONTAINING PROTEIN (PTHR14191) |
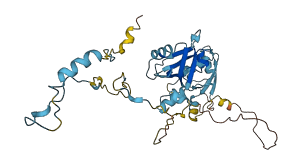
Descriptions
The mammalian Na+/H+ exchange regulatory factor 1 (NHERF1) is a multidomain scaffolding protein essential for regulating the intracellular trafficking and macromolecular assembly of transmembrane ion channels and receptors. NHERF1 is held in an autoinhibited state through intramolecular interactions between PDZ2 and the CT domain that also includes a C-terminal PDZ-binding motif (-SNL). A central feature of NHERF1 is its binding to ezrin and to other ezrin-radixin-moesin (ERM) proteins, forming a communication bridge between plasma membrane proteins and the actin cytoskeleton ezrin modulates the PDZ domains of NHERF1 to assemble multiprotein complexes in a cooperative fashion. Upon binding to ezrin, the intramolecular domain-domain interaction is released.
Autoinhibitory domains (AIDs)
Target domain |
14-94 (PDZ1 domain); 154-234 (PDZ2 domain) |
Relief mechanism |
Partner binding |
Assay |
Structural analysis |
Accessory elements
No accessory elements
Autoinhibited structure
Activated structure

21 structures for O14745
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 1G9O | X-ray | 150 A | A | 11-99 | PDB |
| 1GQ4 | X-ray | 190 A | A | 11-94 | PDB |
| 1GQ5 | X-ray | 220 A | A | 11-94 | PDB |
| 1I92 | X-ray | 170 A | A | 11-94 | PDB |
| 1SGH | X-ray | 350 A | B | 321-358 | PDB |
| 2D10 | X-ray | 250 A | E/F/G/H | 331-358 | PDB |
| 2JXO | NMR | - | A | 150-240 | PDB |
| 2KJD | NMR | - | A | 150-270 | PDB |
| 2KRG | NMR | - | A | 150-358 | PDB |
| 2M0T | NMR | - | A | 11-120 | PDB |
| 2M0U | NMR | - | A | 11-120 | PDB |
| 2M0V | NMR | - | A | 150-270 | PDB |
| 2OZF | X-ray | 150 A | A | 150-235 | PDB |
| 4JL7 | X-ray | 116 A | A | 11-95 | PDB |
| 4LMM | X-ray | 110 A | A | 11-94 | PDB |
| 4MPA | X-ray | 110 A | A | 11-94 | PDB |
| 4N6X | X-ray | 105 A | A | 11-94 | PDB |
| 4PQW | X-ray | 147 A | A | 11-94 | PDB |
| 4Q3H | X-ray | 144 A | A/B | 150-234 | PDB |
| 6RQR | X-ray | 220 A | A/B | 150-269 | PDB |
| AF-O14745-F1 | Predicted | AlphaFoldDB |
372 variants for O14745
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs35910969 CA117368 RCV000005588 |
110 | L>V | Nephrolithiasis/osteoporosis, hypophosphatemic, 2 (nphlop2) Hypophosphatemic nephrolithiasis/osteoporosis 2 [Ensembl, ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA117369 RCV000005589 rs41282065 RCV000484640 |
153 | R>Q | Nephrolithiasis/osteoporosis, hypophosphatemic, 2 (nphlop2) Hypophosphatemic nephrolithiasis/osteoporosis 2 [Ensembl, ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
CA8750680 rs147104235 RCV000680101 |
219 | I>M | Hypophosphatemic nephrolithiasis/osteoporosis 2 [ClinVar] | Yes |
ClinGen ClinVar ESP ExAC TOPMed dbSNP gnomAD |
|
CA117370 RCV000005590 RCV002054417 RCV000454997 RCV000662330 rs119486097 |
225 | E>K | Nephrolithiasis/osteoporosis, hypophosphatemic, 2 (nphlop2) Hypophosphatemic nephrolithiasis/osteoporosis 2 [Ensembl, ClinVar] | Yes |
ClinGen ClinVar 1000Genomes ESP TOPMed dbSNP gnomAD |
|
CA400935767 rs1324850593 |
2 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
CA400935771 rs1400553918 |
2 | S>R | No |
ClinGen gnomAD |
|
|
CA8750519 rs769816246 |
3 | A>G | No |
ClinGen ExAC gnomAD |
|
|
CA400935776 rs769816246 |
3 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA400935782 rs1371847700 |
4 | D>V | No |
ClinGen gnomAD |
|
|
rs1223666496 CA400935787 |
5 | A>T | No |
ClinGen gnomAD |
|
|
rs759634038 CA8750521 |
5 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs1212202572 CA400935800 |
7 | A>V | No |
ClinGen gnomAD |
|
|
CA8750522 rs772345283 |
8 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772345283 CA400935805 |
8 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1307479058 CA400935802 |
8 | G>R | No |
ClinGen TOPMed |
|
|
CA8750523 rs775644483 |
9 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA293924373 rs942896360 |
10 | P>R | No |
ClinGen TOPMed |
|
|
rs777026648 CA8750525 |
12 | P>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400935822 rs777026648 |
12 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1163868389 CA400935843 |
15 | C>F | No |
ClinGen gnomAD |
|
|
rs1400579001 CA400935855 |
17 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
CA8750527 rs201821766 |
18 | E>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1326302401 CA400935874 |
20 | G>S | No |
ClinGen gnomAD |
|
|
rs1353463791 CA400935882 |
21 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1448079674 CA400935885 |
21 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1448079674 CA400935884 |
21 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1353463791 CA400935880 |
21 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1357908170 CA400935896 |
23 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400935899 rs1231865609 |
23 | G>V | No |
ClinGen gnomAD |
|
|
rs1292995957 CA400935901 |
24 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1216134173 CA400935903 |
24 | Y>S | No |
ClinGen TOPMed |
|
|
rs1216500303 CA400935909 |
25 | G>R | No |
ClinGen TOPMed gnomAD |
|
|
CA400935908 rs1216500303 |
25 | G>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1273007489 CA400935915 |
26 | F>L | No |
ClinGen TOPMed |
|
|
rs757966579 CA293924452 |
27 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750530 rs765897805 |
29 | H>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs903284621 CA400935939 |
29 | H>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs751217204 CA8750531 |
30 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400935943 rs1390982901 |
30 | G>R | No |
ClinGen TOPMed |
|
|
CA400935957 rs1403241286 |
32 | K>T | No |
ClinGen gnomAD |
|
|
CA8750532 rs754627534 |
33 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750533 rs780880396 |
34 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750535 rs780010556 |
34 | K>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400935971 rs1346430700 |
34 | K>R | No |
ClinGen gnomAD |
|
|
CA8750538 rs374560863 |
37 | Q>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1377809441 CA400935995 |
38 | Y>D | No |
ClinGen gnomAD |
|
|
rs1312680441 CA400936001 |
39 | I>L | No |
ClinGen gnomAD |
|
|
CA293924491 rs1033435287 |
40 | R>Q | No |
ClinGen gnomAD |
|
|
CA400936009 rs1341577481 |
40 | R>W | No |
ClinGen gnomAD |
|
|
CA8750539 rs368955507 |
41 | L>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400936026 rs1418096580 |
43 | E>A | No |
ClinGen TOPMed |
|
|
CA400936029 rs1219891231 |
43 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
CA400936028 rs1219891231 |
43 | E>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1338968839 CA400936022 |
43 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1338968839 CA400936023 |
43 | E>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs35738271 CA8750540 |
44 | P>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs747239085 CA8750541 |
44 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs35738271 CA400936030 |
44 | P>S | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1289723190 CA400936037 |
45 | G>D | No |
ClinGen gnomAD |
|
|
CA8750543 rs776990193 |
46 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400936042 rs1489999120 |
46 | S>P | No |
ClinGen gnomAD |
|
|
CA400936049 rs1241173893 |
47 | P>L | No |
ClinGen gnomAD |
|
|
CA400936047 rs1190735666 |
47 | P>S | No |
ClinGen gnomAD |
|
|
rs765579547 CA400936058 |
49 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs966154702 CA293924594 |
49 | E>G | No |
ClinGen gnomAD |
|
|
rs765579547 CA8750545 |
49 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400936068 rs1433540629 |
50 | K>R | No |
ClinGen gnomAD |
|
|
CA8750546 rs773765376 |
51 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1315317808 CA400936074 |
51 | A>V | No |
ClinGen TOPMed |
|
|
CA8750548 rs765846960 |
52 | G>E | No |
ClinGen ExAC gnomAD |
|
|
CA8750547 rs577901501 |
52 | G>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA400936116 rs1305870256 |
55 | A>V | No |
ClinGen gnomAD |
|
|
CA293924636 rs200659767 |
56 | G>R | No |
ClinGen 1000Genomes gnomAD |
|
|
rs751038948 CA8750549 |
57 | D>G | No |
ClinGen ExAC gnomAD |
|
|
rs1352675686 CA400936136 |
57 | D>Y | No |
ClinGen gnomAD |
|
|
CA293924649 rs372160939 |
58 | R>Q | No |
ClinGen ESP gnomAD |
|
|
rs368325587 CA8750550 |
58 | R>W | No |
ClinGen ESP ExAC gnomAD |
|
|
rs201382493 CA8750551 |
60 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs752546414 CA8750552 |
61 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs777700723 CA8750554 |
62 | V>G | No |
ClinGen ExAC gnomAD |
|
|
rs756041118 CA8750553 |
62 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA400936217 rs1364893506 |
63 | N>D | No |
ClinGen gnomAD |
|
|
CA8750555 rs376770068 |
66 | N>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA400936293 rs1157211822 |
67 | V>M | No |
ClinGen gnomAD |
|
|
RCV000722205 rs1567820191 |
68 | E>missing | No |
ClinVar dbSNP |
|
|
rs139622189 RCV000521667 CA8750557 |
68 | E>A | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA400936350 rs1328250383 |
70 | E>K | No |
ClinGen gnomAD |
|
|
CA400936392 rs1337503157 |
72 | H>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs768761604 CA8750559 |
72 | H>R | No |
ClinGen ExAC gnomAD |
|
|
CA400936382 rs1378088620 |
72 | H>Y | No |
ClinGen TOPMed |
|
|
CA8750560 rs776902657 |
73 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400936397 rs776902657 |
73 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750561 rs748352647 |
73 | Q>L | No |
ClinGen ExAC gnomAD |
|
|
CA293924787 rs1025981329 |
78 | R>C | No |
ClinGen Ensembl |
|
|
CA400936485 rs914885580 |
80 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs914885580 CA293924793 |
80 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs914885580 CA400936484 |
80 | R>S | No |
ClinGen TOPMed gnomAD |
|
|
CA8750563 rs201874131 |
81 | A>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1198761510 CA400936496 |
82 | A>S | No |
ClinGen gnomAD |
|
|
CA400936499 rs1251988791 |
82 | A>V | No |
ClinGen gnomAD |
|
|
CA8750564 rs763400849 |
83 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1363720161 CA400936506 |
84 | N>D | No |
ClinGen gnomAD |
|
|
rs781350887 CA293924816 |
84 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1485048608 CA400936510 |
84 | N>S | No |
ClinGen TOPMed |
|
|
rs1347256478 CA400936512 |
85 | A>T | No |
ClinGen TOPMed |
|
|
CA293924825 rs908869976 |
86 | V>L | No |
ClinGen Ensembl |
|
|
rs934163535 CA293924829 |
87 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs959095474 CA293924834 |
87 | R>H | No |
ClinGen gnomAD |
|
|
CA400936543 rs1398795274 |
91 | V>I | No |
ClinGen gnomAD |
|
|
CA400936549 rs1328178314 |
92 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs760384895 CA8750571 |
94 | E>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400936563 rs760384895 |
94 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400936579 rs1218915402 |
96 | D>G | No |
ClinGen gnomAD |
|
|
CA8750572 rs764023307 |
96 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1252668447 CA400936587 |
97 | E>G | No |
ClinGen gnomAD |
|
|
rs1434899856 CA400936583 |
97 | E>K | No |
ClinGen TOPMed |
|
|
rs1479408450 CA400936595 |
98 | Q>R | No |
ClinGen gnomAD |
|
|
CA8750573 rs753650408 |
103 | G>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400936627 rs753650408 |
103 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1038411133 CA293924925 |
106 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
CA400936650 rs1357979063 |
107 | R>G | No |
ClinGen gnomAD |
|
|
CA8750574 rs537924938 |
109 | E>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA400936667 rs1167350392 |
109 | E>D | No |
ClinGen gnomAD |
|
|
rs556165171 CA8750575 |
109 | E>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1354626148 CA400936683 |
112 | R>L | No |
ClinGen gnomAD |
|
|
CA293925001 rs1007573140 |
113 | A>P | No |
ClinGen TOPMed gnomAD |
|
|
rs1305339091 CA400936693 |
114 | Q>R | No |
ClinGen gnomAD |
|
|
rs1485442722 CA400936731 |
120 | A>T | No |
ClinGen TOPMed |
|
|
CA8750577 rs781225507 |
120 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1349267248 CA400936736 |
121 | E>K | No |
ClinGen gnomAD |
|
|
CA400936745 rs1213706099 |
122 | P>A | No |
ClinGen gnomAD |
|
|
CA400936748 rs528033874 |
122 | P>L | No |
ClinGen 1000Genomes TOPMed |
|
|
CA293925047 rs528033874 |
122 | P>R | No |
ClinGen 1000Genomes TOPMed |
|
|
CA400936752 rs1186623830 |
123 | P>L | No |
ClinGen gnomAD |
|
|
CA400936751 rs1485589998 |
123 | P>S | No |
ClinGen gnomAD |
|
|
CA400936767 rs1426895066 |
126 | A>T | No |
ClinGen gnomAD |
|
|
rs1192603284 CA400936772 |
126 | A>V | No |
ClinGen gnomAD |
|
|
rs1395819936 CA400936780 |
127 | E>D | No |
ClinGen gnomAD |
|
|
CA400936786 rs1269662949 |
128 | V>E | No |
ClinGen Ensembl |
|
|
CA400936785 rs1269662949 |
128 | V>G | No |
ClinGen Ensembl |
|
|
CA400936781 rs1385946003 |
128 | V>L | No |
ClinGen gnomAD |
|
|
rs1171761541 CA400936795 |
130 | G>R | No |
ClinGen gnomAD |
|
|
CA400936801 rs1374757474 |
131 | A>T | No |
ClinGen gnomAD |
|
|
rs1598336792 CA400936822 |
134 | E>K | No |
ClinGen Ensembl |
|
|
CA400936839 rs1454129192 |
136 | E>* | No |
ClinGen gnomAD |
|
|
CA400936865 rs1290342520 |
139 | E>D | No |
ClinGen gnomAD |
|
|
CA400936869 rs1354404293 |
140 | A>D | No |
ClinGen TOPMed |
|
|
CA293925111 rs1003334664 |
142 | K>N | No |
ClinGen TOPMed gnomAD |
|
|
CA400936902 rs1381259383 |
142 | K>R | No |
ClinGen gnomAD |
|
|
rs1411150897 CA400936918 |
143 | S>N | No |
ClinGen TOPMed |
|
|
rs778131619 CA8750581 |
145 | P>Q | No |
ClinGen ExAC |
|
|
rs1026436067 CA293925131 |
146 | E>G | No |
ClinGen gnomAD |
|
|
rs1231440104 CA400936967 |
146 | E>K | No |
ClinGen gnomAD |
|
|
rs1231440104 CA400936963 |
146 | E>Q | No |
ClinGen gnomAD |
|
|
CA293925144 rs950926703 |
147 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1598342937 CA400938763 |
148 | R>C | No |
ClinGen Ensembl |
|
|
rs370781758 CA8750596 |
148 | R>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs370781758 CA400938765 |
148 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750599 rs756341850 |
149 | E>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750598 rs752725349 |
149 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs777906037 CA400938789 |
150 | L>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs777906037 CA8750600 |
150 | L>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs771386787 CA8750602 |
151 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs541355230 CA8750601 |
151 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750603 rs779127116 |
153 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs772669485 CA8750604 |
155 | C>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750605 rs775166247 |
156 | T>A | No |
ClinGen ExAC gnomAD |
|
|
rs1371737560 CA400938843 |
156 | T>I | No |
ClinGen gnomAD |
|
|
CA293940063 rs967768641 |
157 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
CA400938884 rs1294246004 |
159 | K>T | No |
ClinGen gnomAD |
|
|
rs1337248315 CA400938911 |
161 | P>L | No |
ClinGen gnomAD |
|
|
CA8750606 rs746464571 |
163 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400938945 rs1470649814 |
164 | Y>C | No |
ClinGen gnomAD |
|
|
CA400938989 rs749814524 |
167 | N>I | No |
ClinGen ExAC gnomAD |
|
|
CA8750608 rs749814524 |
167 | N>S | No |
ClinGen ExAC gnomAD |
|
|
CA400938986 rs749814524 |
167 | N>T | No |
ClinGen ExAC gnomAD |
|
|
CA293940125 rs373243340 |
168 | L>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs373243340 CA8750610 |
168 | L>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750611 rs772840570 |
169 | H>P | No |
ClinGen ExAC gnomAD |
|
|
CA400939046 rs1193436828 |
171 | D>E | No |
ClinGen TOPMed |
|
|
rs762899974 CA8750612 |
171 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA8750613 RCV000887508 rs149161808 |
173 | S>F | No |
ClinGen ClinVar 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
|
CA8750614 rs752778784 |
174 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1163841803 CA400939107 |
176 | G>D | No |
ClinGen gnomAD |
|
|
rs1163841803 CA400939110 |
176 | G>V | No |
ClinGen gnomAD |
|
|
CA8750615 rs756254122 |
177 | Q>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400939114 rs756254122 |
177 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400939140 rs1598343026 |
179 | I>L | No |
ClinGen Ensembl |
|
|
rs146832150 CA8750617 |
180 | R>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750618 rs146832150 |
180 | R>P | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs146832150 CA8750619 |
180 | R>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs200609318 CA8750616 |
180 | R>W | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs573918301 CA8750620 |
182 | V>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1353296873 CA400939170 |
182 | V>M | No |
ClinGen gnomAD |
|
|
CA8750623 rs200926419 |
185 | D>E | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs768327472 CA400939234 |
186 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs768327472 CA8750624 |
186 | S>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775971197 CA8750625 |
187 | P>L | No |
ClinGen ExAC gnomAD |
|
|
COSM3820503 rs775971197 CA8750626 |
187 | P>R | breast [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA8750628 rs773071671 |
189 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA400939289 rs1157237860 |
191 | S>* | No |
ClinGen Ensembl |
|
|
rs774331136 CA400939319 |
194 | R>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774331136 CA8750631 |
194 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766158601 CA8750630 |
194 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA400939334 rs1293007256 |
195 | A>V | No |
ClinGen TOPMed |
|
|
rs956472744 CA293940298 |
196 | Q>L | No |
ClinGen Ensembl |
|
|
CA400939361 rs1242342663 |
197 | D>V | No |
ClinGen gnomAD |
|
|
rs764277046 CA8750633 |
198 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs764277046 CA400939370 |
198 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400939366 rs1476164667 |
198 | R>S | No |
ClinGen gnomAD |
|
|
rs753824169 CA8750634 |
199 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs765322250 CA8750636 |
200 | V>A | No |
ClinGen ExAC gnomAD |
|
|
rs757524454 CA8750635 |
200 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400939399 rs1377786866 |
201 | E>K | No |
ClinGen TOPMed gnomAD |
|
|
rs778705591 CA8750667 |
204 | G>R | No |
ClinGen ExAC gnomAD |
|
|
CA8750669 rs772055107 |
207 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs745595834 CA8750668 |
207 | M>T | No |
ClinGen ExAC |
|
|
CA400940007 rs1198830308 |
208 | E>K | No |
ClinGen gnomAD |
|
|
rs760635571 CA293941688 |
209 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750671 rs760635571 |
209 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750673 rs773383515 |
212 | H>L | No |
ClinGen ExAC gnomAD |
|
|
CA8750675 rs766649573 |
215 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750677 rs751975880 |
217 | S>P | No |
ClinGen ExAC |
|
|
rs1329530680 CA400940074 |
218 | A>D | No |
ClinGen gnomAD |
|
|
rs551375430 CA8750679 |
218 | A>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs756614733 CA8750682 |
220 | R>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA293941752 rs1005570601 |
220 | R>S | No |
ClinGen Ensembl |
|
|
CA400940093 rs1335101430 |
221 | A>V | No |
ClinGen gnomAD |
|
|
CA8750683 rs777298818 |
222 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs373197801 CA8750686 |
223 | G>E | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs756919776 CA8750685 |
223 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400940107 rs1440018823 |
224 | D>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1598343796 CA400940146 |
230 | V>G | No |
ClinGen Ensembl |
|
|
CA400940143 rs1457482619 |
230 | V>L | No |
ClinGen gnomAD |
|
|
CA400940151 rs1598343803 |
231 | V>G | No |
ClinGen Ensembl |
|
|
CA8750694 rs749312548 |
231 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400940149 rs749312548 |
231 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400940157 rs774771671 |
232 | D>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750696 rs774771671 |
232 | D>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1014461649 CA293941867 |
233 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
rs1020861597 CA293941874 |
233 | R>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1416389395 CA400940179 |
234 | E>K | No |
ClinGen gnomAD |
|
|
RCV000054713 CA216337 rs387907536 |
236 | D>E | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs1291121264 CA400940212 |
236 | D>N | No |
ClinGen gnomAD |
|
|
CA8750699 rs369327875 |
237 | E>* | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750698 rs369327875 |
237 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750700 rs369327875 |
237 | E>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA293941932 rs373061379 |
241 | K>R | No |
ClinGen ESP gnomAD |
|
|
rs201989899 CA8750702 |
242 | C>Y | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA400940349 rs1190440315 |
243 | R>K | No |
ClinGen gnomAD |
|
|
CA400940352 rs1190440315 |
243 | R>T | No |
ClinGen gnomAD |
|
|
rs756831725 CA8750703 |
244 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1423425353 CA400940423 |
248 | Q>K | No |
ClinGen gnomAD |
|
|
rs764717359 CA8750704 |
250 | H>N | No |
ClinGen ExAC gnomAD |
|
|
CA293945231 rs1027498511 |
257 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA400941213 rs1428096974 |
258 | P>L | No |
ClinGen TOPMed |
|
|
rs575858499 CA8750726 |
258 | P>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA8750728 rs754888522 |
261 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM984118 CA293945283 rs754888522 |
261 | N>S | endometrium [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs781103116 CA8750729 |
262 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs756065565 CA8750731 |
263 | E>K | No |
ClinGen ExAC gnomAD |
|
|
CA400941350 rs1462034651 |
264 | I>T | No |
ClinGen TOPMed |
|
|
rs1424550335 CA400941365 |
265 | Q>* | No |
ClinGen gnomAD |
|
|
CA8750779 rs755209522 |
267 | E>K | No |
ClinGen ExAC |
|
|
CA400941666 rs1174791941 |
268 | N>H | No |
ClinGen gnomAD |
|
|
rs781453513 CA293946795 |
268 | N>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750781 rs755665225 |
269 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750782 rs150507328 |
270 | R>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs777978291 CA8750783 |
270 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs777978291 CA8750784 |
270 | R>L | No |
ClinGen ExAC gnomAD |
|
|
rs777978291 CA400941712 |
270 | R>P | No |
ClinGen ExAC gnomAD |
|
|
CA8750785 rs771302473 |
271 | E>D | No |
ClinGen ExAC gnomAD |
|
|
CA293946823 rs774000677 |
274 | A>E | No |
ClinGen ExAC gnomAD |
|
|
rs1318580314 CA400941752 |
274 | A>S | No |
ClinGen gnomAD |
|
|
CA8750786 rs774000677 |
274 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA8750787 rs745316613 |
275 | E>Q | No |
ClinGen ExAC gnomAD |
|
|
CA400941778 rs1384257631 |
276 | A>S | No |
ClinGen TOPMed |
|
|
CA400941775 rs1384257631 |
276 | A>T | No |
ClinGen TOPMed |
|
|
CA400941783 rs1320432550 |
276 | A>V | No |
ClinGen TOPMed |
|
|
CA400941796 rs1342676283 |
277 | A>V | No |
ClinGen gnomAD |
|
|
CA8750788 rs771691982 |
278 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750789 rs775346495 |
279 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs775346495 CA400941821 |
279 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400941827 rs1477455500 |
280 | S>C | No |
ClinGen TOPMed |
|
|
rs192227522 CA8750790 |
280 | S>N | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750791 rs763994159 |
281 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA400941842 rs1598346069 |
281 | P>S | No |
ClinGen Ensembl |
|
|
rs1468515434 CA400941850 |
282 | R>G | No |
ClinGen gnomAD |
|
|
rs1200282377 CA400941854 |
282 | R>K | No |
ClinGen gnomAD |
|
|
CA400941872 rs1365707474 |
283 | P>L | No |
ClinGen Ensembl |
|
|
rs1413019076 CA400941917 |
287 | R>S | No |
ClinGen gnomAD |
|
|
rs1229030372 CA400941927 |
288 | S>C | No |
ClinGen gnomAD |
|
|
rs1388081112 CA400941932 |
289 | A>T | No |
ClinGen gnomAD |
|
|
CA400941942 rs1441710219 |
289 | A>V | No |
ClinGen gnomAD |
|
|
CA400941944 rs1428891804 |
290 | S>P | No |
ClinGen TOPMed |
|
|
CA293946880 rs1020242568 |
293 | T>A | No |
ClinGen TOPMed |
|
|
rs765104894 CA8750795 |
294 | S>I | No |
ClinGen ExAC gnomAD |
|
|
rs765104894 CA400941996 |
294 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs755119543 CA8750797 |
295 | E>G | No |
ClinGen ExAC gnomAD |
|
|
rs750349847 CA8750796 |
295 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767555645 CA8750798 |
296 | E>G | No |
ClinGen ExAC gnomAD |
|
|
CA400942095 rs1176169810 |
298 | N>K | No |
ClinGen gnomAD |
|
|
CA400942112 rs1567826304 |
300 | Q>* | No |
ClinGen Ensembl |
|
|
CA400942124 rs1366836783 |
301 | D>H | No |
ClinGen gnomAD |
|
|
rs141613848 CA8750824 |
301 | D>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA293947352 rs569186312 |
303 | P>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8750825 rs569186312 |
303 | P>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA8750826 rs375430336 |
304 | P>A | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1307140220 CA400942165 |
304 | P>R | No |
ClinGen gnomAD |
|
|
rs938666218 CA293947375 |
305 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
CA400942168 rs938666218 |
305 | K>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA8750827 rs758923316 |
305 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs371164739 CA8750828 |
306 | Q>R | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750829 rs746450148 |
307 | D>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750830 rs768201403 |
307 | D>V | No |
ClinGen ExAC gnomAD |
|
|
rs1298994306 CA400942200 |
308 | S>C | No |
ClinGen gnomAD |
|
|
rs1344882857 CA400942204 |
309 | T>P | No |
ClinGen TOPMed gnomAD |
|
|
CA8750831 rs375193779 |
310 | A>V | No |
ClinGen ESP ExAC gnomAD |
|
|
rs769628266 CA8750834 |
311 | P>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750833 rs769628266 |
311 | P>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs762910299 CA8750835 |
312 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA400942230 rs1567826349 |
314 | T>A | No |
ClinGen Ensembl |
|
|
CA8750839 rs760721877 |
315 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA8750838 rs146249672 |
315 | S>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA8750841 rs754043982 |
316 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA8750846 rs780488061 |
318 | D>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750844 rs750773487 |
318 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs750773487 CA8750845 |
318 | D>Y | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1300723847 CA400942266 |
319 | P>A | No |
ClinGen gnomAD |
|
|
rs387907537 CA216339 RCV000054714 |
320 | I>F | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs752105483 CA8750848 |
320 | I>M | No |
ClinGen ExAC gnomAD |
|
|
CA400942282 rs1472959207 |
320 | I>N | No |
ClinGen TOPMed |
|
|
rs1405098726 CA400942304 |
322 | D>E | No |
ClinGen gnomAD |
|
|
CA400942330 rs1411379865 |
324 | N>S | No |
ClinGen TOPMed |
|
|
CA400942340 rs1362370010 |
325 | I>L | No |
ClinGen TOPMed gnomAD |
|
|
rs780732081 CA8750850 |
325 | I>S | No |
ClinGen ExAC gnomAD |
|
|
CA8750851 rs780732081 |
325 | I>T | No |
ClinGen ExAC gnomAD |
|
|
rs1362370010 CA400942342 |
325 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
CA400942360 rs1468089649 |
326 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1295493783 CA400942353 |
326 | S>P | No |
ClinGen gnomAD |
|
|
rs1221311881 CA400942380 |
328 | A>V | No |
ClinGen TOPMed |
|
|
rs1209468952 CA400942404 |
329 | M>I | No |
ClinGen gnomAD |
|
|
CA400942410 rs1286950048 |
330 | A>T | No |
ClinGen TOPMed |
|
|
rs777549830 CA8750854 |
330 | A>V | No |
ClinGen ExAC gnomAD |
|
|
rs770699023 CA8750856 |
332 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs749024706 CA8750855 |
332 | E>V | No |
ClinGen ExAC gnomAD |
|
|
rs534067895 CA8750857 |
333 | R>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1173646134 CA400942498 |
334 | A>V | No |
ClinGen gnomAD |
|
|
rs1457486939 CA400942514 |
335 | H>P | No |
ClinGen gnomAD |
|
|
CA400942537 rs1160884660 |
336 | Q>R | No |
ClinGen gnomAD |
|
|
CA8750860 rs768665440 |
338 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA8750861 rs776719627 |
338 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1359713263 CA400942590 |
339 | S>N | No |
ClinGen gnomAD |
|
|
CA8750862 rs762153640 |
340 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA293947672 rs957974780 |
341 | K>* | No |
ClinGen TOPMed |
|
|
CA400942630 rs1299582805 |
341 | K>R | No |
ClinGen gnomAD |
|
|
CA8750864 rs773559482 |
342 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA293947680 rs554217161 |
342 | R>W | No |
ClinGen 1000Genomes TOPMed |
|
|
rs387907535 RCV000054711 CA216333 |
344 | P>A | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs199563744 CA8750865 |
344 | P>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs199563744 CA400942673 |
344 | P>Q | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs199563744 CA8750866 |
344 | P>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA293947714 rs963229518 |
346 | M>I | No |
ClinGen Ensembl |
|
|
CA293947726 rs200658339 |
347 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750869 rs200658339 |
347 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA8750870 rs144154644 |
348 | W>* | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1489393185 CA400942749 |
348 | W>* | No |
ClinGen gnomAD |
|
|
CA400942751 rs1489393185 |
348 | W>C | No |
ClinGen gnomAD |
|
|
CA400942792 rs1265356095 |
351 | K>R | No |
ClinGen TOPMed gnomAD |
|
|
rs144091666 CA8750872 COSM277349 |
353 | E>K | large_intestine [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA293947771 rs951159583 |
354 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1409241993 CA400942827 |
356 | S>N | No |
ClinGen gnomAD |
No associated diseases with O14745
1 regional properties for O14745
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Ets domain | 4 - 90 | IPR000418 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR14191 | PDZ DOMAIN CONTAINING PROTEIN |
| PANTHER Subfamily | PTHR14191:SF7 | NA(+)_H(+) EXCHANGE REGULATORY COFACTOR NHE-RF1 |
| PANTHER Protein Class | scaffold/adaptor protein | |
| PANTHER Pathway Category | No pathway information available | |
18 GO annotations of cellular component
| Name | Definition |
|---|---|
| actin cytoskeleton | The part of the cytoskeleton (the internal framework of a cell) composed of actin and associated proteins. Includes actin cytoskeleton-associated complexes. |
| apical plasma membrane | The region of the plasma membrane located at the apical end of the cell. |
| brush border membrane | The portion of the plasma membrane surrounding the brush border. |
| cell periphery | The part of a cell encompassing the cell cortex, the plasma membrane, and any external encapsulating structures. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| endomembrane system | A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| filopodium | Thin, stiff, actin-based protrusion extended by the leading edge of a motile cell such as a crawling fibroblast or amoeba, or an axonal or dendritic growth cone, or a dendritic shaft. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| microvillus | Thin cylindrical membrane-covered projections on the surface of an animal cell containing a core bundle of actin filaments. Present in especially large numbers on the absorptive surface of intestinal cells. |
| microvillus membrane | The portion of the plasma membrane surrounding a microvillus. |
| perinuclear region of cytoplasm | Cytoplasm situated near, or occurring around, the nucleus. |
| plasma membrane protein complex | Any protein complex that is part of the plasma membrane. |
| ruffle | Projection at the leading edge of a crawling cell; the protrusions are supported by a microfilament meshwork. |
| sperm midpiece | The highly organized segment of the sperm flagellum which begins at the connecting piece and is characterized by the presence of 9 outer dense fibers (ODFs) that lie outside each of the 9 outer axonemal microtubule doublets and by a sheath of mitochondria that encloses the ODFs and the axoneme; the midpiece terminates about one-fourth of the way down the sperm flagellum at the annulus, which marks the beginning of the principal piece. |
| stereocilium tip | A distinct compartment at the tip of a stereocilium, distal to the site of attachment to the apical cell surface. It consists of a dense matrix bridging the barbed ends of the stereocilium actin filaments with the overlying plasma membrane, is dynamic compared to the shaft, and is required for stereocilium elongation. |
| vesicle | Any small, fluid-filled, spherical organelle enclosed by membrane. |
16 GO annotations of molecular function
| Name | Definition |
|---|---|
| beta-2 adrenergic receptor binding | Binding to a beta-2 adrenergic receptor. |
| beta-catenin binding | Binding to a catenin beta subunit. |
| chloride channel regulator activity | Binds to and modulates the activity of a chloride channel. |
| dopamine receptor binding | Binding to a dopamine receptor. |
| gamma-aminobutyric acid transmembrane transporter activity | Enables the transfer of gamma-aminobutyric acid from one side of a membrane to the other. Gamma-aminobutyric acid is 4-aminobutyrate (GABA). |
| growth factor receptor binding | Binding to a growth factor receptor. |
| myosin II binding | Binding to a class II myosin, any member of the class of 'conventional' double-headed myosins that includes muscle myosin. |
| PDZ domain binding | Binding to a PDZ domain of a protein, a domain found in diverse signaling proteins. |
| phosphatase binding | Binding to a phosphatase. |
| protein N-terminus binding | Binding to a protein N-terminus, the end of any peptide chain at which the 2-amino (or 2-imino) function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein self-association | Binding to a domain within the same polypeptide. |
| protein-containing complex binding | Binding to a macromolecular complex. |
| protein-membrane adaptor activity | The binding activity of a molecule that brings together a protein or a protein complex with a membrane, or bringing together two membranes, either via membrane lipid binding or by interacting with a membrane protein, to establish or maintain the localization of the protein, protein complex or organelle. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
| type 2 metabotropic glutamate receptor binding | Binding to a type 2 metabotropic glutamate receptor. |
| type 3 metabotropic glutamate receptor binding | Binding to a type 3 metabotropic glutamate receptor. |
43 GO annotations of biological process
| Name | Definition |
|---|---|
| actin cytoskeleton organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of cytoskeletal structures comprising actin filaments and their associated proteins. |
| adenylate cyclase-activating dopamine receptor signaling pathway | An adenylate cyclase-activating G protein-coupled receptor signaling pathway initiated by dopamine binding to its receptor, and ending with the regulation of a downstream cellular process. |
| auditory receptor cell stereocilium organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a stereocilium. A stereocilium is an actin-based protrusion from the apical surface of auditory hair cells. |
| bile acid secretion | The regulated release of bile acid, composed of any of a group of steroid carboxylic acids occurring in bile, by a cell or a tissue. |
| cellular phosphate ion homeostasis | Any process involved in the maintenance of an internal steady state of phosphate ions at the level of a cell. |
| cerebrospinal fluid circulation | The neurological system process driven by motile cilia on ependymal cells of the brain by which cerebrospinal fluid circulates from the sites of secretion to the sites of absorption. In ventricular cavities, the flow is unidirectional and rostrocaudal, in subarachnoid spaces, the flow is multi-directional. |
| cilium organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of a cilium, a specialized eukaryotic organelle that consists of a filiform extrusion of the cell surface. Each cilium is bounded by an extrusion of the cytoplasmic membrane, and contains a regular longitudinal array of microtubules, anchored basally in a centriole. |
| establishment of epithelial cell apical/basal polarity | The specification and formation of the apicobasal polarity of an epithelial cell. |
| establishment of Golgi localization | The directed movement of the Golgi to a specific location. |
| gamma-aminobutyric acid import | The directed movement of gamma-aminobutyric acid (GABA, 4-aminobutyrate) into a cell or organelle. |
| gland morphogenesis | The process in which the anatomical structures of a gland are generated and organized. |
| glutathione transport | The directed movement of glutathione, the tripeptide glutamylcysteinylglycine, into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| import across plasma membrane | The directed movement of some substance from outside of a cell, across the plasma membrane and into the cytosol. |
| maintenance of epithelial cell apical/basal polarity | The maintenance of the apicobasal polarity of an epithelial cell. |
| microvillus assembly | Formation of a microvillus, a thin cylindrical membrane-covered projection on the surface of a cell. |
| morphogenesis of an epithelium | The process in which the anatomical structures of epithelia are generated and organized. An epithelium consists of closely packed cells arranged in one or more layers, that covers the outer surfaces of the body or lines any internal cavity or tube. |
| negative regulation of canonical Wnt signaling pathway | Any process that decreases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| negative regulation of cell migration | Any process that stops, prevents, or reduces the frequency, rate or extent of cell migration. |
| negative regulation of cell population proliferation | Any process that stops, prevents or reduces the rate or extent of cell proliferation. |
| negative regulation of ERK1 and ERK2 cascade | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| negative regulation of mitotic cell cycle | Any process that stops, prevents or reduces the rate or extent of progression through the mitotic cell cycle. |
| negative regulation of phosphatidylinositol 3-kinase signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of signal transduction mediated by the phosphatidylinositol 3-kinase cascade. |
| negative regulation of platelet-derived growth factor receptor signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of the platelet-derived growth factor receptor signaling pathway. |
| negative regulation of protein kinase B signaling | Any process that stops, prevents, or reduces the frequency, rate or extent of protein kinase B signaling, a series of reactions mediated by the intracellular serine/threonine kinase protein kinase B. |
| negative regulation of sodium:proton antiporter activity | Any process that stops or reduces the activity of a sodium:hydrogen antiporter, which catalyzes the reaction: Na+(out) + H+(in) = Na+(in) + H+(out). |
| nuclear migration | The directed movement of the nucleus to a specific location within a cell. |
| phospholipase C-activating dopamine receptor signaling pathway | A phospholipase C-activating receptor G protein-coupled receptor signaling pathway initiated by dopamine binding to its receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| plasma membrane organization | A process that is carried out at the cellular level which results in the assembly, arrangement of constituent parts, or disassembly of the plasma membrane. |
| positive regulation of intrinsic apoptotic signaling pathway | Any process that activates or increases the frequency, rate or extent of intrinsic apoptotic signaling pathway. |
| positive regulation of ion transmembrane transport | Any process that activates or increases the frequency, rate or extent of the directed movement of ions from one side of a membrane to the other. |
| protein localization to plasma membrane | A process in which a protein is transported to, or maintained in, a specific location in the plasma membrane. |
| protein-containing complex assembly | The aggregation, arrangement and bonding together of a set of macromolecules to form a protein-containing complex. |
| regulation of cell shape | Any process that modulates the surface configuration of a cell. |
| regulation of cell size | Any process that modulates the size of a cell. |
| regulation of excretion | Any process that modulates the frequency, rate, or extent of excretion, the elimination by an organism of the waste products that arise as a result of metabolic activity. |
| regulation of protein kinase activity | Any process that modulates the frequency, rate or extent of protein kinase activity. |
| regulation of sodium:proton antiporter activity | Any process that modulates the activity of a sodium:hydrogen antiporter, which catalyzes the reaction: Na+(out) + H+(in) = Na+(in) + H+(out). |
| renal absorption | A renal system process in which water, ions, glucose and proteins are taken up from the collecting ducts, glomerulus and proximal and distal loops of the nephron. In non-mammalian species, absorption may occur in related structures (e.g. protein absorption is observed in nephrocytes in Drosophila, see PMID:23264686). |
| renal phosphate ion absorption | A renal system process in which phosphate ions are taken up from the collecting ducts and proximal and distal loops of the nephron. In non-mammalian species, absorption may occur in related structures. |
| renal sodium ion transport | The directed movement of sodium ions (Na+) by the renal system. |
| sensory perception of sound | The series of events required for an organism to receive an auditory stimulus, convert it to a molecular signal, and recognize and characterize the signal. Sonic stimuli are detected in the form of vibrations and are processed to form a sound. |
| transport across blood-brain barrier | The directed movement of substances (e.g. macromolecules, small molecules, ions) through the blood-brain barrier. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
11 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3SZK8 | NHERF1 | Na | Bos taurus (Bovine) | SS |
| Q5ZM14 | NHERF1 | Na | Gallus gallus (Chicken) | SS |
| Q24008 | inaD | Inactivation-no-after-potential D protein | Drosophila melanogaster (Fruit fly) | PR |
| Q9Y6N9 | USH1C | Harmonin | Homo sapiens (Human) | PR |
| Q86UT5 | PDZD3 | Na(+)/H(+) exchange regulatory cofactor NHE-RF4 | Homo sapiens (Human) | PR |
| Q99MJ6 | Pdzd3 | Na(+)/H(+) exchange regulatory cofactor NHE-RF4 | Mus musculus (Mouse) | PR |
| Q9JIL4 | Pdzk1 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | Mus musculus (Mouse) | PR |
| Q0QWG9 | Grid2ip | Delphilin | Mus musculus (Mouse) | PR |
| P70441 | Slc9a3r1 | Na(+)/H(+) exchange regulatory cofactor NHE-RF1 | Mus musculus (Mouse) | PR |
| Q9JJ40 | Pdzk1 | Na(+)/H(+) exchange regulatory cofactor NHE-RF3 | Rattus norvegicus (Rat) | PR |
| Q9JJ19 | Nherf1 | Na | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSADAAAGAP | LPRLCCLEKG | PNGYGFHLHG | EKGKLGQYIR | LVEPGSPAEK | AGLLAGDRLV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EVNGENVEKE | THQQVVSRIR | AALNAVRLLV | VDPETDEQLQ | KLGVQVREEL | LRAQEAPGQA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| EPPAAAEVQG | AGNENEPREA | DKSHPEQREL | RPRLCTMKKG | PSGYGFNLHS | DKSKPGQFIR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SVDPDSPAEA | SGLRAQDRIV | EVNGVCMEGK | QHGDVVSAIR | AGGDETKLLV | VDRETDEFFK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| KCRVIPSQEH | LNGPLPVPFT | NGEIQKENSR | EALAEAALES | PRPALVRSAS | SDTSEELNSQ |
| 310 | 320 | 330 | 340 | 350 | |
| DSPPKQDSTA | PSSTSSSDPI | LDFNISLAMA | KERAHQKRSS | KRAPQMDWSK | KNELFSNL |