O14733
Gene name |
MAP2K7 (JNKK2, MEK7, MKK7, PRKMK7, SKK4) |
Protein name |
Dual specificity mitogen-activated protein kinase kinase 7 |
Names |
MAP kinase kinase 7, MAPKK 7, JNK-activating kinase 2, MAPK/ERK kinase 7, MEK 7, Stress-activated protein kinase kinase 4, SAPK kinase 4, SAPKK-4, SAPKK4, c-Jun N-terminal kinase kinase 2, JNK kinase 2, JNKK 2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:5609 |
EC number |
2.7.12.2: Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
Protein Class |
|
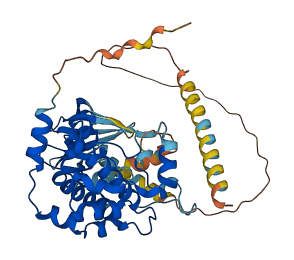
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
260-282 (Activation loop from InterPro)
Target domain |
120-380 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

37 structures for O14733
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2DYL | X-ray | 245 A | A | 101-405 | PDB |
| 3WZU | X-ray | 301 A | A | 103-419 | PDB |
| 4UX9 | X-ray | 234 A | F/G/H/I | 37-48 | PDB |
| 5B2K | X-ray | 275 A | A | 103-419 | PDB |
| 5B2L | X-ray | 210 A | A | 103-419 | PDB |
| 5B2M | X-ray | 306 A | A | 103-419 | PDB |
| 5Y8U | X-ray | 292 A | A | 103-419 | PDB |
| 5Y90 | X-ray | 130 A | A | 103-419 | PDB |
| 5Z1D | X-ray | 228 A | A | 102-419 | PDB |
| 5Z1E | X-ray | 230 A | A | 102-419 | PDB |
| 6IB0 | X-ray | 260 A | A | 101-408 | PDB |
| 6IB2 | X-ray | 210 A | A | 101-408 | PDB |
| 6QFL | X-ray | 220 A | A | 101-408 | PDB |
| 6QFR | X-ray | 230 A | A | 101-408 | PDB |
| 6QFT | X-ray | 270 A | A | 101-408 | PDB |
| 6QG4 | X-ray | 230 A | A | 111-408 | PDB |
| 6QG7 | X-ray | 210 A | A | 101-408 | PDB |
| 6QHO | X-ray | 270 A | A | 111-408 | PDB |
| 6QHR | X-ray | 252 A | A | 111-408 | PDB |
| 6YFZ | X-ray | 190 A | A | 100-405 | PDB |
| 6YG0 | X-ray | 200 A | A | 100-405 | PDB |
| 6YG1 | X-ray | 222 A | A/B/C | 60-405 | PDB |
| 6YG2 | X-ray | 200 A | A | 100-405 | PDB |
| 6YG3 | X-ray | 205 A | A | 100-405 | PDB |
| 6YG4 | X-ray | 230 A | A | 100-405 | PDB |
| 6YG5 | X-ray | 240 A | A | 100-405 | PDB |
| 6YG6 | X-ray | 215 A | A/B | 100-405 | PDB |
| 6YG7 | X-ray | 220 A | A/B | 100-405 | PDB |
| 6YZ4 | X-ray | 170 A | A | 100-405 | PDB |
| 7CBX | X-ray | 206 A | A | 103-419 | PDB |
| 7OVI | X-ray | 195 A | A | 101-408 | PDB |
| 7OVJ | X-ray | 235 A | A | 101-408 | PDB |
| 7OVK | X-ray | 205 A | A | 101-408 | PDB |
| 7OVL | X-ray | 290 A | A | 101-408 | PDB |
| 7OVM | X-ray | 290 A | A | 101-408 | PDB |
| 7OVN | X-ray | 290 A | A | 101-408 | PDB |
| AF-O14733-F1 | Predicted | AlphaFoldDB |
297 variants for O14733
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000505552 CA403679635 rs1555701191 |
251 | L>P | Neuroblastoma [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA9148223 rs763754700 |
2 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1443440676 CA403677842 |
6 | L>V | No |
ClinGen gnomAD |
|
|
COSM1397681 rs1301547539 CA403677856 |
8 | Q>* | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA403677886 rs1433064449 |
12 | R>L | No |
ClinGen TOPMed |
|
|
CA9148229 rs766283924 |
22 | R>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9148228 rs750181141 |
22 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs751559509 CA9148230 |
25 | R>W | No |
ClinGen ExAC gnomAD |
|
|
rs1403159799 CA403677981 |
27 | R>M | No |
ClinGen gnomAD |
|
|
CA403678016 rs1169050667 |
32 | L>P | No |
ClinGen gnomAD |
|
|
CA403678014 rs1453514771 |
32 | L>V | No |
ClinGen gnomAD |
|
|
rs749341799 CA9148233 |
33 | D>E | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 33 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9148234 rs757454138 |
34 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1460437314 CA403678026 |
34 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1391474983 CA403678044 |
36 | P>L | No |
ClinGen gnomAD |
|
|
rs1370547916 CA403678055 |
38 | R>Q | No |
ClinGen gnomAD |
|
|
CA403678054 rs1324055434 |
38 | R>W | No |
ClinGen gnomAD |
|
|
CA9148235 rs779037374 |
40 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs533132248 CA304961967 |
42 | T>A | No |
ClinGen 1000Genomes TOPMed |
|
|
CA304967969 rs1024708183 |
44 | Q>R | No |
ClinGen TOPMed |
|
|
CA304967970 rs967433524 |
46 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs967433524 CA403678224 |
46 | P>R | No |
ClinGen TOPMed gnomAD |
|
|
rs778375800 CA9148308 |
50 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs957493807 CA304967979 |
51 | G>A | No |
ClinGen TOPMed |
|
|
CA403678251 rs1284071014 |
51 | G>R | No |
ClinGen gnomAD |
|
|
CA403678253 rs1284071014 |
51 | G>W | No |
ClinGen gnomAD |
|
|
rs748767169 CA9148309 |
52 | G>A | No |
ClinGen ExAC gnomAD |
|
|
rs1484368982 CA403678262 |
53 | S>G | No |
ClinGen gnomAD |
|
|
CA403678264 rs1186294216 |
53 | S>N | No |
ClinGen gnomAD |
|
|
CA9148310 rs756811414 |
54 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs533357050 CA9148311 |
54 | R>H | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs745585391 CA9148312 |
55 | S>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA304967997 rs976387571 |
57 | S>F | No |
ClinGen TOPMed |
|
|
rs1435136254 CA403678294 |
58 | S>L | No |
ClinGen gnomAD |
|
|
rs1175363004 CA403678316 |
61 | S>F | No |
ClinGen gnomAD |
|
|
rs775089795 CA9148314 |
62 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA403678319 rs1373620825 |
62 | P>S | No |
ClinGen gnomAD |
|
|
rs1337733940 CA403678326 |
63 | Q>R | No |
ClinGen gnomAD |
|
|
CA403678334 rs1252128134 |
64 | H>R | No |
ClinGen TOPMed |
|
|
rs1440233859 CA403678343 |
65 | P>L | No |
ClinGen gnomAD |
|
|
rs1213372895 CA403678339 |
65 | P>S | No |
ClinGen TOPMed |
|
|
rs746839170 CA9148315 |
66 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1308848837 CA403678344 |
66 | T>P | No |
ClinGen gnomAD |
|
|
rs1044083059 CA304968056 |
67 | P>R | No |
ClinGen gnomAD |
|
|
CA304968032 rs948274560 |
67 | P>S | No |
ClinGen Ensembl |
|
|
CA403678355 rs1347873574 |
68 | P>S | No |
ClinGen gnomAD |
|
| TCGA novel | 69 | A>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1316384071 CA403678364 |
69 | A>V | No |
ClinGen TOPMed |
|
|
CA403678367 rs1439065076 |
70 | R>P | No |
ClinGen gnomAD |
|
|
CA403678368 rs1439065076 |
70 | R>Q | No |
ClinGen gnomAD |
|
|
rs768434615 CA9148316 |
70 | R>W | No |
ClinGen ExAC gnomAD |
|
|
CA403678376 rs1237190214 |
72 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs758493704 CA9148317 COSM3693225 |
72 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
CA403678396 rs1195341922 |
74 | M>I | No |
ClinGen gnomAD |
|
|
CA304968081 rs138677713 |
75 | L>M | No |
ClinGen Ensembl |
|
|
rs1393160324 CA403678406 |
76 | G>E | No |
ClinGen gnomAD |
|
| TCGA novel | 76 | G>W | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1168449280 CA403678409 |
77 | L>F | No |
ClinGen TOPMed gnomAD |
|
|
CA403678424 rs1408284809 |
79 | S>L | No |
ClinGen gnomAD |
|
|
CA403678441 rs1372070521 |
82 | F>C | No |
ClinGen gnomAD |
|
|
CA403678442 rs1452479193 |
82 | F>L | No |
ClinGen TOPMed |
|
|
CA403678444 rs763099575 |
83 | T>A | No |
ClinGen ExAC TOPMed |
|
|
CA9148318 rs763099575 |
83 | T>P | No |
ClinGen ExAC TOPMed |
|
|
CA403678454 rs1599625307 |
84 | P>R | No |
ClinGen Ensembl |
|
|
rs766635456 CA9148319 |
85 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1301746163 CA403678457 |
85 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
rs766635456 CA403678455 |
85 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1469619336 CA403678461 |
86 | S>G | No |
ClinGen TOPMed |
|
|
rs1331007401 CA403678463 |
86 | S>N | No |
ClinGen gnomAD |
|
|
rs774579720 CA9148320 |
87 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs1476247422 CA403678569 |
98 | E>D | No |
ClinGen gnomAD |
|
|
rs1177582341 CA403678579 |
100 | M>V | No |
ClinGen gnomAD |
|
|
COSM3773231 CA9148337 rs769867905 |
103 | T>M | Variant assessed as Somatic; 0.0 impact. pancreas urinary_tract [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs759693800 CA9148339 |
104 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs1383410204 CA403678624 |
106 | L>P | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
| TCGA novel | 107 | T>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403678630 rs1256880340 |
107 | T>S | No |
ClinGen TOPMed |
|
|
CA403678633 rs1314051512 |
108 | I>V | No |
ClinGen gnomAD |
|
|
CA9148342 rs760936521 CA304968429 |
109 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148343 rs557240660 |
110 | G>D | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA403678643 rs1310113723 |
110 | G>S | No |
ClinGen gnomAD |
|
| TCGA novel | 112 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403678671 rs1191058688 |
112 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
CA403678676 rs1428581952 |
113 | Y>H | No |
ClinGen gnomAD |
|
| TCGA novel | 114 | Q>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1323652275 CA403678694 |
115 | A>G | No |
ClinGen TOPMed |
|
|
CA304968685 rs28395770 |
116 | E>K | No |
ClinGen Ensembl |
|
|
rs979747164 CA304968687 |
118 | N>D | No |
ClinGen Ensembl |
|
|
CA9148389 rs56316660 VAR_040825 |
118 | N>S | No |
ClinGen UniProt ESP ExAC TOPMed dbSNP gnomAD |
|
|
rs1326208752 CA403678722 |
119 | D>E | No |
ClinGen gnomAD |
|
|
rs1464734841 CA403678716 |
119 | D>N | No |
ClinGen gnomAD |
|
|
CA9148392 rs753585786 |
123 | L>F | No |
ClinGen ExAC gnomAD |
|
|
rs372177955 CA9148391 |
123 | L>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1292855446 CA403678752 |
124 | G>C | No |
ClinGen gnomAD |
|
| TCGA novel | 125 | E>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
COSM568986 CA403678790 rs1276435407 |
129 | G>D | lung Variant assessed as Somatic; 0.0 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA9148396 rs755099146 |
132 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
| TCGA novel | 134 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
| TCGA novel | 135 | W>* | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9148397 rs56106612 VAR_040826 |
138 | R>C | No |
ClinGen UniProt 1000Genomes ExAC TOPMed dbSNP gnomAD |
|
|
rs748434331 CA9148398 |
138 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA403678854 rs748434331 |
138 | R>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148400 rs773578983 |
139 | F>L | No |
ClinGen ExAC gnomAD |
|
|
rs771439651 CA9148402 |
140 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749648216 CA9148401 |
140 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763308668 CA304968795 |
141 | K>E | No |
ClinGen TOPMed gnomAD |
|
|
rs376991761 CA9148403 |
141 | K>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1205135126 CA403678876 |
142 | T>I | No |
ClinGen TOPMed |
|
|
CA9148405 rs771779482 |
143 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 144 | H>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403678885 rs1233328050 |
144 | H>Y | No |
ClinGen TOPMed |
|
|
CA403678894 rs1401718147 |
145 | V>G | No |
ClinGen gnomAD |
|
|
CA304968823 rs1049003237 |
145 | V>I | No |
ClinGen TOPMed gnomAD |
|
|
rs200200323 CA9148407 |
146 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs761549769 CA9148410 |
148 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1045580606 CA304968973 |
152 | R>Q | No |
ClinGen TOPMed gnomAD |
|
|
rs761655517 CA9148427 COSM3693227 |
152 | R>W | large_intestine [Cosmic] | No |
ClinGen cosmic curated ExAC gnomAD |
|
CA403678956 COSM1680582 rs1441304667 |
153 | R>H | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA403678968 rs1568278051 |
155 | G>E | No |
ClinGen Ensembl |
|
|
rs1384098610 CA403678980 |
157 | K>E | No |
ClinGen gnomAD |
|
|
CA9148429 rs367833467 |
157 | K>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
| TCGA novel | 159 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403679014 rs1174269918 |
161 | K>R | No |
ClinGen TOPMed gnomAD |
|
| VAR_040827 | 162 | R>C | a colorectal adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
| VAR_040828 | 162 | R>H | a colorectal adenocarcinoma sample; somatic mutation [UniProt] | No | UniProt |
|
rs528497143 CA9148430 |
164 | L>F | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1413990204 CA403679045 |
166 | D>N | No |
ClinGen gnomAD |
|
|
CA9148433 rs756198072 |
169 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1372204318 CA403679100 |
174 | H>Y | No |
ClinGen gnomAD |
|
|
CA403679112 rs370478429 |
175 | D>E | No |
ClinGen ESP ExAC gnomAD |
|
|
rs754078664 CA9148435 |
175 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA9148438 rs746254265 |
177 | P>L | No |
ClinGen ExAC gnomAD |
|
|
rs779365415 CA9148437 |
177 | P>S | No |
ClinGen ExAC gnomAD |
|
|
rs975881933 CA304969056 |
178 | Y>C | No |
ClinGen Ensembl |
|
| TCGA novel | 178 | Y>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1196504228 CA403679133 |
179 | I>V | No |
ClinGen gnomAD |
|
|
COSM1680583 rs746502176 CA9148441 |
180 | V>M | pancreas Variant assessed as Somatic; impact. haematopoietic_and_lymphoid_tissue [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA403679164 rs1375915089 |
183 | F>S | No |
ClinGen gnomAD |
|
|
rs776342808 CA9148443 |
185 | T>M | No |
ClinGen ExAC gnomAD |
|
|
CA403679178 rs776342808 |
185 | T>R | No |
ClinGen ExAC gnomAD |
|
|
rs769499056 CA9148445 |
188 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA304969089 rs887570441 |
189 | N>D | No |
ClinGen gnomAD |
|
|
CA9148446 rs199553372 |
189 | N>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9148488 COSM1397683 rs377648179 |
190 | T>M | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA403679224 rs1160351321 |
191 | D>N | No |
ClinGen TOPMed |
|
|
CA304972245 rs55800262 VAR_040829 |
195 | A>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen UniProt Ensembl NCI-TCGA dbSNP |
|
CA403679263 rs1440693094 |
196 | M>I | No |
ClinGen gnomAD |
|
|
rs775543935 CA9148493 |
199 | M>T | No |
ClinGen ExAC gnomAD |
|
|
CA403679302 rs1433998983 |
202 | C>Y | No |
ClinGen TOPMed |
|
|
rs1174847870 CA403679307 |
203 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA403679313 rs1467142899 |
204 | E>K | No |
ClinGen gnomAD |
|
|
rs773428876 CA9148496 |
205 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1401142748 CA403679330 |
206 | L>F | No |
ClinGen gnomAD |
|
|
CA403679341 rs1568278464 |
207 | K>N | No |
ClinGen Ensembl |
|
|
CA9148497 rs763209807 |
208 | K>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148499 rs751828868 |
209 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148498 rs552080947 |
209 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9148500 rs759996040 |
210 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA403679354 rs1301903022 |
210 | M>L | No |
ClinGen gnomAD |
|
|
rs753313895 CA9148502 |
211 | Q>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148501 rs767899243 |
211 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs756619690 CA9148503 |
212 | G>D | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 214 | I>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403679384 rs1192222025 |
215 | P>T | No |
ClinGen gnomAD |
|
| TCGA novel | 216 | E>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs745624779 CA9148508 |
217 | R>C | No |
ClinGen ExAC gnomAD |
|
|
rs868265912 CA304972375 |
217 | R>H | Variant assessed as Somatic; 4.813e-05 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA403679401 rs868265912 |
217 | R>L | No |
ClinGen gnomAD |
|
|
rs771955180 CA9148509 |
218 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs372316207 CA304972405 |
221 | K>R | No |
ClinGen TOPMed |
|
|
rs373789571 CA304972409 |
224 | V>E | No |
ClinGen ESP TOPMed |
|
|
rs746875567 CA9148517 |
225 | A>V | Variant assessed as Somatic; 9.758e-05 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1421560106 CA403679470 |
227 | V>M | No |
ClinGen gnomAD |
|
|
rs765979119 CA9148557 |
228 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs1383490584 CA403679485 |
229 | A>S | No |
ClinGen TOPMed |
|
|
CA403679487 rs1325957309 |
229 | A>V | No |
ClinGen gnomAD |
|
|
rs1377356152 CA403679498 |
231 | Y>C | No |
ClinGen gnomAD |
|
|
CA403679506 rs1310627873 |
232 | Y>C | No |
ClinGen gnomAD |
|
|
CA403679518 rs1356525990 |
234 | K>T | No |
ClinGen gnomAD |
|
|
CA403679537 rs748034368 |
236 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs906866888 CA304972862 |
237 | H>L | No |
ClinGen TOPMed |
|
|
CA403679544 rs756020181 |
237 | H>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1283210992 CA403679541 |
237 | H>Y | No |
ClinGen gnomAD |
|
|
rs1210306090 CA403679546 |
238 | G>S | No |
ClinGen gnomAD |
|
| TCGA novel | 239 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403679559 rs1489727215 |
240 | I>V | No |
ClinGen gnomAD |
|
|
CA403679567 rs1277929944 |
241 | H>Y | No |
ClinGen TOPMed |
|
|
rs1156815343 COSM1165620 CA403679580 |
243 | D>N | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs769071544 CA9148568 |
244 | V>I | No |
ClinGen ExAC gnomAD |
|
|
rs769071544 CA403679587 |
244 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1338503438 CA403679621 |
249 | I>V | No |
ClinGen TOPMed |
|
|
CA403679627 rs1599627776 |
250 | L>M | No |
ClinGen Ensembl |
|
|
rs1409519936 CA403679632 |
251 | L>M | No |
ClinGen gnomAD |
|
|
CA9148572 rs772735492 |
252 | D>G | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 252 | D>H | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1238524673 CA403679644 |
253 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs1599627826 CA403679649 |
253 | E>V | No |
ClinGen Ensembl |
|
|
CA304972896 rs913866951 |
254 | R>Q | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA403679653 rs1279498308 |
254 | R>W | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA403679665 rs1302728142 |
256 | Q>L | No |
ClinGen gnomAD |
|
|
CA403679666 rs1302728142 |
256 | Q>R | No |
ClinGen gnomAD |
|
|
CA403679672 rs1332085514 |
257 | I>F | No |
ClinGen gnomAD |
|
|
CA403679673 rs1439010437 |
257 | I>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1439010437 CA403679674 |
257 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA403679671 rs1332085514 |
257 | I>V | No |
ClinGen gnomAD |
|
|
rs1053566 CA304972901 VAR_029890 |
259 | L>F | No |
ClinGen UniProt Ensembl dbSNP |
|
|
rs1599627883 CA403679690 |
260 | C>S | No |
ClinGen Ensembl |
|
|
CA9148578 rs752458822 |
263 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs1474172928 CA403679742 |
267 | R>C | No |
ClinGen TOPMed |
|
|
rs1177861432 CA403679758 |
270 | D>H | No |
ClinGen Ensembl |
|
|
CA403679781 rs1332304138 |
273 | A>S | No |
ClinGen gnomAD |
|
|
CA403679798 rs1445100216 |
275 | T>K | No |
ClinGen gnomAD |
|
|
rs1445100216 CA403679796 COSM302613 |
275 | T>M | ovary Variant assessed as Somatic; 0.0 impact. oesophagus central_nervous_system [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA403679800 rs1332216223 |
276 | R>W | No |
ClinGen gnomAD |
|
|
CA304972946 rs942074295 |
282 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
| rs757758116 | 286 | P>= | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1190484047 CA403679895 |
288 | R>C | No |
ClinGen gnomAD |
|
|
CA9148678 rs779562628 |
288 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs1312792372 CA403679918 |
291 | P>L | No |
ClinGen gnomAD |
|
|
rs779379219 CA9148681 |
294 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs746557259 CA9148682 |
295 | T>P | No |
ClinGen ExAC gnomAD |
|
|
rs768160589 CA9148683 |
296 | K>N | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 296 | K>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1341085574 CA403679956 |
298 | D>N | No |
ClinGen TOPMed |
|
|
COSM1641275 rs1425924011 CA403679969 |
299 | Y>C | Variant assessed as Somatic; 0.0 impact. stomach [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA403679979 rs1599628435 |
300 | D>E | No |
ClinGen Ensembl |
|
|
COSM3938384 rs1185925179 CA403679989 |
302 | R>Q | Variant assessed as Somatic; 0.0 impact. oesophagus [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
CA9148686 rs769642939 |
304 | D>N | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs879150050 CA304973305 |
305 | V>I | No |
ClinGen Ensembl |
|
|
CA304973328 rs1046459698 COSM94662 |
311 | S>L | lung [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA403680143 rs1369799132 |
317 | T>I | No |
ClinGen gnomAD |
|
| TCGA novel | 322 | Y>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs778579797 CA9148731 |
331 | V>I | No |
ClinGen ExAC gnomAD |
|
|
CA403680268 rs1568279607 |
335 | V>I | No |
ClinGen Ensembl |
|
|
CA403680301 rs1417048704 |
339 | E>D | No |
ClinGen gnomAD |
|
| TCGA novel | 341 | P>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA304973526 rs982599133 |
342 | L>F | No |
ClinGen TOPMed gnomAD |
|
| TCGA novel | 343 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403680326 rs1302574218 |
344 | P>S | No |
ClinGen gnomAD |
|
|
CA304973544 rs752181956 |
345 | G>A | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148743 rs752181956 |
345 | G>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148742 rs200517538 COSM1003407 |
345 | G>R | endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
CA304973546 rs915467733 |
346 | H>Y | No |
ClinGen TOPMed |
|
|
CA403680366 rs1351336395 |
350 | S>L | No |
ClinGen TOPMed |
|
|
CA9148746 rs753467019 |
351 | G>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA304973568 rs918854402 |
354 | Q>H | No |
ClinGen gnomAD |
|
|
CA403680394 rs1222436068 |
354 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
CA9148748 rs778725144 |
355 | S>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9148750 rs758376970 |
357 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
rs1182280031 CA403680432 |
360 | C>R | No |
ClinGen gnomAD |
|
|
CA403680502 rs1334645480 |
367 | K>N | No |
ClinGen gnomAD |
|
| TCGA novel | 368 | R>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
rs1330906396 CA403680512 |
369 | P>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
CA403680526 rs1405096609 |
371 | Y>D | No |
ClinGen gnomAD |
|
| TCGA novel | 373 | K>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403680543 rs1284773326 |
373 | K>T | No |
ClinGen TOPMed gnomAD |
|
|
rs1432947387 CA403680555 |
375 | L>F | No |
ClinGen gnomAD |
|
|
CA304974473 rs923590375 |
378 | S>N | No |
ClinGen TOPMed |
|
|
CA403680591 rs923590375 |
378 | S>T | No |
ClinGen TOPMed |
|
|
rs1312906351 CA403680604 |
380 | I>F | No |
ClinGen gnomAD |
|
|
CA9148821 rs772810212 |
380 | I>N | No |
ClinGen ExAC gnomAD |
|
|
rs774212941 CA9148824 |
382 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA9148825 rs759229847 |
382 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA9148826 rs372974513 |
384 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA403680640 rs1431888473 |
385 | T>M | No |
ClinGen TOPMed |
|
|
rs764054804 CA403680641 |
386 | L>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1331773120 CA403680643 |
386 | L>P | No |
ClinGen TOPMed |
|
| TCGA novel | 386 | L>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA403680646 rs1267005014 |
387 | E>K | No |
ClinGen gnomAD |
|
|
CA403680664 rs1159130971 |
389 | D>G | No |
ClinGen gnomAD |
|
|
rs1451089097 CA403680669 |
390 | V>L | No |
ClinGen gnomAD |
|
|
CA403680679 rs1406524174 |
391 | A>V | No |
ClinGen gnomAD |
|
|
CA403680703 rs1464023661 |
395 | K>E | No |
ClinGen TOPMed |
|
|
rs1334570087 CA403680707 |
395 | K>R | No |
ClinGen gnomAD |
|
|
rs1391918745 CA403680729 |
398 | M>T | No |
ClinGen gnomAD |
|
|
rs201368825 CA9148834 COSM1397688 |
399 | A>V | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
CA9148836 rs202245509 |
404 | P>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
rs556127943 CA9148840 |
405 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs778482297 CA9148839 |
405 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1599630114 CA403680778 |
406 | T>I | No |
ClinGen Ensembl |
|
|
CA9148841 rs377699571 |
406 | T>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA403680790 rs1161324664 |
408 | G>D | No |
ClinGen gnomAD |
|
|
rs745460007 CA9148843 |
408 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA403680796 rs1173924867 |
409 | V>A | No |
ClinGen gnomAD |
|
|
CA9148845 rs775112625 COSM1397689 |
409 | V>I | Variant assessed as Somatic; 0.0 impact. large_intestine [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
| TCGA novel | 410 | L>P | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9148846 rs760564022 |
411 | S>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA403680816 rs1458046756 |
412 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
rs764009958 CA9148847 |
413 | P>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA304974649 rs533495400 |
413 | P>T | No |
ClinGen Ensembl |
|
|
CA403680828 rs761745379 |
414 | H>Q | No |
ClinGen ExAC gnomAD |
|
|
CA9148848 rs776620038 |
414 | H>Y | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
COSM1527476 rs1599630216 CA403680839 |
416 | P>L | lung [Cosmic] | No |
ClinGen cosmic curated Ensembl |
|
CA403680844 rs1221943389 |
417 | F>S | No |
ClinGen gnomAD |
|
|
CA403680849 rs1359556850 |
418 | F>L | No |
ClinGen gnomAD |
|
|
rs1055882048 CA304974670 |
419 | R>G | No |
ClinGen TOPMed |
No associated diseases with O14733
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.12.2 | Dual-specificity kinases (those acting on Ser/Thr and Tyr residues) |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
13 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| enzyme binding | Binding to an enzyme, a protein with catalytic activity. |
| JUN kinase kinase activity | Catalysis of the phosphorylation of tyrosine and threonine residues in a c-Jun NH2-terminal kinase (JNK), a member of a subgroup of mitogen-activated protein kinases (MAPKs), which signal in response to cytokines and exposure to environmental stress. JUN kinase kinase (JNKK) is a dual-specificity protein kinase kinase and requires activation by a serine/threonine kinase JUN kinase kinase kinase. |
| magnesium ion binding | Binding to a magnesium (Mg) ion. |
| MAP kinase activity | Catalysis of the reaction: protein + ATP = protein phosphate + ADP. This reaction is the phosphorylation of proteins. Mitogen-activated protein kinase; a family of protein kinases that perform a crucial step in relaying signals from the plasma membrane to the nucleus. They are activated by a wide range of proliferation- or differentiation-inducing signals; activation is strong with agonists such as polypeptide growth factors and tumor-promoting phorbol esters, but weak (in most cell backgrounds) by stress stimuli. |
| MAP kinase kinase activity | Catalysis of the concomitant phosphorylation of threonine (T) and tyrosine (Y) residues in a Thr-Glu-Tyr (TEY) thiolester sequence in a MAP kinase (MAPK) substrate. |
| mitogen-activated protein kinase kinase kinase binding | Binding to a mitogen-activated protein kinase kinase kinase, a protein that can phosphorylate a MAP kinase kinase. |
| molecular function activator activity | A molecular function regulator that activates or increases the activity of its target via non-covalent binding that does not result in covalent modification to the target. |
| protein C-terminus binding | Binding to a protein C-terminus, the end of a peptide chain at which the 1-carboxyl function of a constituent amino acid is not attached in peptide linkage to another amino-acid residue. |
| protein kinase binding | Binding to a protein kinase, any enzyme that catalyzes the transfer of a phosphate group, usually from ATP, to a protein substrate. |
| protein phosphatase binding | Binding to a protein phosphatase. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
24 GO annotations of biological process
| Name | Definition |
|---|---|
| apoptotic process | A programmed cell death process which begins when a cell receives an internal (e.g. DNA damage) or external signal (e.g. an extracellular death ligand), and proceeds through a series of biochemical events (signaling pathway phase) which trigger an execution phase. The execution phase is the last step of an apoptotic process, and is typically characterized by rounding-up of the cell, retraction of pseudopodes, reduction of cellular volume (pyknosis), chromatin condensation, nuclear fragmentation (karyorrhexis), plasma membrane blebbing and fragmentation of the cell into apoptotic bodies. When the execution phase is completed, the cell has died. |
| cellular response to interleukin-1 | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an interleukin-1 stimulus. |
| cellular response to lipopolysaccharide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a lipopolysaccharide stimulus; lipopolysaccharide is a major component of the cell wall of gram-negative bacteria. |
| cellular response to sorbitol | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a sorbitol stimulus. |
| cellular senescence | A cell aging process stimulated in response to cellular stress, whereby normal cells lose the ability to divide through irreversible cell cycle arrest. |
| Fc-epsilon receptor signaling pathway | The series of molecular signals initiated by the binding of the Fc portion of immunoglobulin E (IgE) to an Fc-epsilon receptor on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. The Fc portion of an immunoglobulin is its C-terminal constant region. |
| JNK cascade | An intracellular protein kinase cascade containing at least a JNK (a MAPK), a JNKK (a MAPKK) and a JUN3K (a MAP3K). The cascade can also contain an additional tier: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinases in the downstream tier to transmit a signal within a cell. |
| MAPK cascade | An intracellular protein kinase cascade containing at least a MAPK, a MAPKK and a MAP3K. The cascade can also contain an additional tiers: the upstream MAP4K. The kinases in each tier phosphorylate and activate the kinase in the downstream tier to transmit a signal within a cell. |
| positive regulation of DNA-templated transcription | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of ERK1 and ERK2 cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| positive regulation of JNK cascade | Any process that activates or increases the frequency, rate or extent of signal transduction mediated by the JNK cascade. |
| positive regulation of JUN kinase activity | Any process that activates or increases the frequency, rate or extent of JUN kinase activity. |
| positive regulation of neuron apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death of neurons by apoptotic process. |
| positive regulation of telomerase activity | Any process that activates or increases the frequency, rate or extent of telomerase activity, the catalysis of the reaction: deoxynucleoside triphosphate + DNA(n) = diphosphate + DNA(n+1). |
| positive regulation of telomere capping | Any process that activates or increases the frequency, rate or extent of telomere capping. |
| positive regulation of telomere maintenance via telomerase | Any process that activates or increases the frequency, rate or extent of the addition of telomeric repeats by telomerase. |
| regulation of motor neuron apoptotic process | Any process that modulates the frequency, rate or extent of motor neuron apoptotic process. |
| response to heat | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a heat stimulus, a temperature stimulus above the optimal temperature for that organism. |
| response to osmotic stress | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating an increase or decrease in the concentration of solutes outside the organism or cell. |
| response to tumor necrosis factor | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a tumor necrosis factor stimulus. |
| response to UV | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an ultraviolet radiation (UV light) stimulus. Ultraviolet radiation is electromagnetic radiation with a wavelength in the range of 10 to 380 nanometers. |
| response to wounding | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating damage to the organism. |
| signal transduction | The cellular process in which a signal is conveyed to trigger a change in the activity or state of a cell. Signal transduction begins with reception of a signal (e.g. a ligand binding to a receptor or receptor activation by a stimulus such as light), or for signal transduction in the absence of ligand, signal-withdrawal or the activity of a constitutively active receptor. Signal transduction ends with regulation of a downstream cellular process, e.g. regulation of transcription or regulation of a metabolic process. Signal transduction covers signaling from receptors located on the surface of the cell and signaling via molecules located within the cell. For signaling between cells, signal transduction is restricted to events at and within the receiving cell. |
| stress-activated MAPK cascade | The series of molecular signals in which a stress-activated MAP kinase cascade relays a signal; MAP kinase cascades involve at least three protein kinase activities and culminate in the phosphorylation and activation of a MAP kinase. |
16 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5E9X2 | MAP2K6 | Dual specificity mitogen-activated protein kinase kinase 6 | Bos taurus (Bovine) | EV |
| P52564 | MAP2K6 | Dual specificity mitogen-activated protein kinase kinase 6 | Homo sapiens (Human) | EV |
| P46734 | MAP2K3 | Dual specificity mitogen-activated protein kinase kinase 3 | Homo sapiens (Human) | SS |
| P45985 | MAP2K4 | Dual specificity mitogen-activated protein kinase kinase 4 | Homo sapiens (Human) | EV |
| Q13163 | MAP2K5 | Dual specificity mitogen-activated protein kinase kinase 5 | Homo sapiens (Human) | PR |
| P36507 | MAP2K2 | Dual specificity mitogen-activated protein kinase kinase 2 | Homo sapiens (Human) | EV |
| Q02750 | MAP2K1 | Dual specificity mitogen-activated protein kinase kinase 1 | Homo sapiens (Human) | EV |
| Q8CE90 | Map2k7 | Dual specificity mitogen-activated protein kinase kinase 7 | Mus musculus (Mouse) | PR |
| P70236 | Map2k6 | Dual specificity mitogen-activated protein kinase kinase 6 | Mus musculus (Mouse) | EV |
| P47809 | Map2k4 | Dual specificity mitogen-activated protein kinase kinase 4 | Mus musculus (Mouse) | EV |
| O09110 | Map2k3 | Dual specificity mitogen-activated protein kinase kinase 3 | Mus musculus (Mouse) | PR |
| Q4KSH7 | Map2k7 | Dual specificity mitogen-activated protein kinase kinase 7 | Rattus norvegicus (Rat) | PR |
| G5EDT6 | jkk-1 | Dual specificity mitogen-activated protein kinase kinase jkk-1 | Caenorhabditis elegans | PR |
| G5EDF7 | sek-1 | Dual specificity mitogen-activated protein kinase kinase sek-1 | Caenorhabditis elegans | PR |
| Q21307 | mek-1 | Dual specificity mitogen-activated protein kinase kinase mek-1 | Caenorhabditis elegans | PR |
| Q9DGE0 | map2k6 | Dual specificity mitogen-activated protein kinase kinase 6 | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAASSLEQKL | SRLEAKLKQE | NREARRRIDL | NLDISPQRPR | PTLQLPLAND | GGSRSPSSES |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SPQHPTPPAR | PRHMLGLPST | LFTPRSMESI | EIDQKLQEIM | KQTGYLTIGG | QRYQAEINDL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ENLGEMGSGT | CGQVWKMRFR | KTGHVIAVKQ | MRRSGNKEEN | KRILMDLDVV | LKSHDCPYIV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QCFGTFITNT | DVFIAMELMG | TCAEKLKKRM | QGPIPERILG | KMTVAIVKAL | YYLKEKHGVI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| HRDVKPSNIL | LDERGQIKLC | DFGISGRLVD | SKAKTRSAGC | AAYMAPERID | PPDPTKPDYD |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IRADVWSLGI | SLVELATGQF | PYKNCKTDFE | VLTKVLQEEP | PLLPGHMGFS | GDFQSFVKDC |
| 370 | 380 | 390 | 400 | 410 | |
| LTKDHRKRPK | YNKLLEHSFI | KRYETLEVDV | ASWFKDVMAK | TESPRTSGVL | SQPHLPFFR |