O08644
Gene name |
Ephb6 (Cekl) |
Protein name |
Ephrin type-B receptor 6 |
Names |
MEP, Tyrosine-protein kinase-defective receptor EPH-6 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:13848 |
EC number |
|
Protein Class |
|
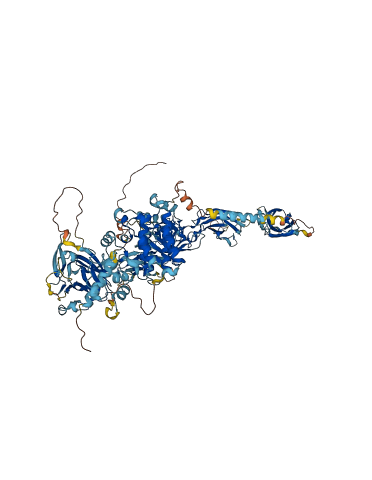
Descriptions
(Annotation based on sequence homology with P29323)
Ephrin type-B receptor 2 is a membrane-associated protein that mediates axon guidance, cell migration and morphogenesis. The Eph receptor tyrosine kinase family is regulated by autophosphorylation within the juxtamembrane region and the kinase activation segment. The structure, supported by mutagenesis data, reveals that the juxtamembrane segment adopts a helical conformation that distorts the small lobe of the kinase domain, and blocks the activation segment from attaining an activated conformation. Phosphorylation of the conserved juxtamembrane tyrosines would relieve this autoinhibition by disturbing the association of the juxtamembrane segment with the kinase domain, while liberating phosphotyrosine sites for binding SH2 domains of target proteins.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for O08644
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O08644-F1 | Predicted | AlphaFoldDB |
38 variants for O08644
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388821707 | 22 | W>* | No | EVA | |
| rs3396855205 | 24 | L>P | No | EVA | |
| rs3396733656 | 25 | V>I | No | EVA | |
| rs3388832191 | 59 | E>G | No | EVA | |
| rs3388818835 | 77 | H>Y | No | EVA | |
| rs3388803415 | 97 | E>D | No | EVA | |
| rs3396266551 | 135 | Q>L | No | EVA | |
| rs3396696297 | 137 | D>H | No | EVA | |
| rs3396648567 | 138 | E>Q | No | EVA | |
| rs3396266563 | 140 | D>E | No | EVA | |
| rs3388815473 | 243 | S>N | No | EVA | |
| rs3388808541 | 255 | G>S | No | EVA | |
| rs37068406 | 325 | P>A | No | EVA | |
| rs3388821737 | 349 | F>S | No | EVA | |
| rs3388818810 | 429 | R>C | No | EVA | |
| rs3388803483 | 430 | Q>* | No | EVA | |
| rs3388818761 | 438 | V>L | No | EVA | |
| rs3388803416 | 516 | L>M | No | EVA | |
| rs3402727888 | 521 | Q>H | No | EVA | |
| rs3388815521 | 531 | M>T | No | EVA | |
| rs3388812713 | 541 | T>I | No | EVA | |
| rs3396679545 | 564 | Y>H | No | EVA | |
| rs3388819482 | 609 | A>V | No | EVA | |
| rs3388825454 | 626 | Q>* | No | EVA | |
| rs3388808590 | 674 | F>Y | No | EVA | |
| rs3388832171 | 688 | R>M | No | EVA | |
| rs3388829246 | 748 | P>S | No | EVA | |
| rs3388818785 | 760 | F>Y | No | EVA | |
| rs3388808588 | 769 | Q>* | No | EVA | |
| rs3388812712 | 797 | N>T | No | EVA | |
| rs3388825165 | 823 | P>S | No | EVA | |
| rs3388832134 | 884 | H>L | No | EVA | |
| rs3388829303 | 885 | L>I | No | EVA | |
| rs3388815551 | 898 | R>Q | No | EVA | |
| rs3388825647 | 904 | L>M | No | EVA | |
| rs3388823546 | 915 | P>S | No | EVA | |
| rs3388793010 | 993 | K>* | No | EVA | |
| rs255736007 | 1005 | H>R | No | EVA |
No associated diseases with O08644
10 regional properties for O08644
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 663 - 912 | IPR000719 |
| domain | Ephrin receptor ligand binding domain | 34 - 232 | IPR001090 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 663 - 906 | IPR001245 |
| conserved_site | Tyrosine-protein kinase, receptor class V, conserved site | 208 - 228 | IPR001426-1 |
| conserved_site | Tyrosine-protein kinase, receptor class V, conserved site | 281 - 301 | IPR001426-2 |
| domain | Sterile alpha motif domain | 938 - 1005 | IPR001660 |
| domain | Fibronectin type III | 364 - 479 | IPR003961-1 |
| domain | Fibronectin type III | 480 - 575 | IPR003961-2 |
| domain | Tyrosine-protein kinase ephrin type A/B receptor-like | 305 - 340 | IPR011641 |
| domain | Ephrin receptor, transmembrane domain | 589 - 660 | IPR027936 |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell surface | The external part of the cell wall and/or plasma membrane. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| neuron projection | A prolongation or process extending from a nerve cell, e.g. an axon or dendrite. |
| receptor complex | Any protein complex that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| transmembrane receptor protein tyrosine kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: ATP + a protein-L-tyrosine = ADP + a protein-L-tyrosine phosphate. |
| transmembrane-ephrin receptor activity | Combining with a transmembrane ephrin to initiate a change in cell activity. |
9 GO annotations of biological process
| Name | Definition |
|---|---|
| activated T cell proliferation | The expansion of a T cell population following activation by an antigenic stimulus. |
| axon guidance | The chemotaxis process that directs the migration of an axon growth cone to a specific target site in response to a combination of attractive and repulsive cues. |
| central nervous system projection neuron axonogenesis | Generation of a long process of a CNS neuron, that carries efferent (outgoing) action potentials from the cell body towards target cells in a different central nervous system region. |
| positive regulation of kinase activity | Any process that activates or increases the frequency, rate or extent of kinase activity, the catalysis of the transfer of a phosphate group, usually from ATP, to a substrate molecule. |
| positive regulation of protein binding | Any process that activates or increases the frequency, rate or extent of protein binding. |
| positive regulation of T cell costimulation | Any process that activates or increases the frequency, rate or extent of T cell costimulation. |
| T cell mediated immunity | Any process involved in the carrying out of an immune response by a T cell. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| type IV hypersensitivity | An inflammatory response driven by T cell recognition of processed soluble or cell-associated antigens leading to cytokine release and leukocyte activation. |
49 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P28693 | EPHB2 | Ephrin type-B receptor 2 | Gallus gallus (Chicken) | PR |
| Q07496 | EPHA4 | Ephrin type-A receptor 4 | Gallus gallus (Chicken) | SS |
| Q07494 | EPHB1 | Ephrin type-B receptor 1 | Gallus gallus (Chicken) | SS |
| Q07497 | EPHB5 | Ephrin type-B receptor 5 | Gallus gallus (Chicken) | PR |
| P29318 | EPHA3 | Ephrin type-A receptor 3 | Gallus gallus (Chicken) | SS |
| O42422 | EPHA7 | Ephrin type-A receptor 7 | Gallus gallus (Chicken) | SS |
| P54755 | EPHA5 | Ephrin type-A receptor 5 | Gallus gallus (Chicken) | SS |
| Q07498 | EPHB3 | Ephrin type-B receptor 3 | Gallus gallus (Chicken) | SS |
| P0C0K6 | EPHB6 | Ephrin type-B receptor 6 | Pan troglodytes (Chimpanzee) | SS |
| P29322 | EPHA8 | Ephrin type-A receptor 8 | Homo sapiens (Human) | SS |
| P21709 | EPHA1 | Ephrin type-A receptor 1 | Homo sapiens (Human) | SS |
| P54764 | EPHA4 | Ephrin type-A receptor 4 | Homo sapiens (Human) | SS |
| P54753 | EPHB3 | Ephrin type-B receptor 3 | Homo sapiens (Human) | SS |
| P29320 | EPHA3 | Ephrin type-A receptor 3 | Homo sapiens (Human) | PR |
| P54760 | EPHB4 | Ephrin type-B receptor 4 | Homo sapiens (Human) | SS |
| P29317 | EPHA2 | Ephrin type-A receptor 2 | Homo sapiens (Human) | SS |
| Q15375 | EPHA7 | Ephrin type-A receptor 7 | Homo sapiens (Human) | SS |
| Q5JZY3 | EPHA10 | Ephrin type-A receptor 10 | Homo sapiens (Human) | SS |
| P29323 | EPHB2 | Ephrin type-B receptor 2 | Homo sapiens (Human) | EV |
| Q9UF33 | EPHA6 | Ephrin type-A receptor 6 | Homo sapiens (Human) | SS |
| P54762 | EPHB1 | Ephrin type-B receptor 1 | Homo sapiens (Human) | SS |
| P54756 | EPHA5 | Ephrin type-A receptor 5 | Homo sapiens (Human) | SS |
| O15197 | EPHB6 | Ephrin type-B receptor 6 | Homo sapiens (Human) | SS |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60629 | Epha5 | Ephrin type-A receptor 5 | Mus musculus (Mouse) | SS |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| P54761 | Ephb4 | Ephrin type-B receptor 4 | Mus musculus (Mouse) | PR |
| Q61772 | Epha7 | Ephrin type-A receptor 7 | Mus musculus (Mouse) | SS |
| Q8CBF3 | Ephb1 | Ephrin type-B receptor 1 | Mus musculus (Mouse) | SS |
| O09127 | Epha8 | Ephrin type-A receptor 8 | Mus musculus (Mouse) | SS |
| Q03137 | Epha4 | Ephrin type-A receptor 4 | Mus musculus (Mouse) | SS |
| P54754 | Ephb3 | Ephrin type-B receptor 3 | Mus musculus (Mouse) | SS |
| Q8BYG9 | Epha10 | Ephrin type-A receptor 10 | Mus musculus (Mouse) | SS |
| P54763 | Ephb2 | Ephrin type-B receptor 2 | Mus musculus (Mouse) | SS |
| Q62413 | Epha6 | Ephrin type-A receptor 6 | Mus musculus (Mouse) | SS |
| P29319 | Epha3 | Ephrin type-A receptor 3 | Mus musculus (Mouse) | SS |
| P54757 | Epha5 | Ephrin type-A receptor 5 | Rattus norvegicus (Rat) | SS |
| P54759 | Epha7 | Ephrin type-A receptor 7 | Rattus norvegicus (Rat) | SS |
| P09759 | Ephb1 | Ephrin type-B receptor 1 | Rattus norvegicus (Rat) | SS |
| O08680 | Epha3 | Ephrin type-A receptor 3 | Rattus norvegicus (Rat) | SS |
| P0C0K7 | Ephb6 | Ephrin type-B receptor 6 | Rattus norvegicus (Rat) | SS |
| O61460 | vab-1 | Ephrin receptor 1 | Caenorhabditis elegans | SS |
| Q8VZJ9 | CRCK2 | Calmodulin-binding receptor-like cytoplasmic kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LMN7 | WAK5 | Wall-associated receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LSV3 | WAKL16 | Putative wall-associated receptor kinase-like 16 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M342 | WAKL15 | Wall-associated receptor kinase-like 15 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O13147 | ephb3 | Ephrin type-B receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O13146 | epha3 | Ephrin type-A receptor 3 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| O73878 | ephb4b | Ephrin type-B receptor 4b | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MATEGTTGSG | SRVVAGMVCS | LWLLVLGSSV | LALEEVLLDT | TGETSEIGWL | TYPPGGWDEV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SVLDDQRRLT | RTFEACHVAG | LPPGSGQDNW | LQTHFVERRG | AQRAHIRLHF | SVRACSSLGV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| SGGTCRETFT | LYYRQADEPD | GPDSIAAWHL | KRWTKVDTIA | ADESFPASSS | SSSWAVGPHR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| TGQRVGLQLN | VKERSFGPLT | QRGFYVAFQD | TGACLALVAV | KLFSYTCPSV | LRAFASFPET |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QASGAGGASL | VAAVGTCVAH | AEPEEDGVGG | QAGGSPPRLH | CNGEGRWMVA | VGGCRCQPGH |
| 310 | 320 | 330 | 340 | 350 | 360 |
| QPARGDKLCQ | ACPEGSYKAL | AGNVPCSPCP | ARSHSPDPAA | PVCPCLQGFY | RASSDPPEAP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| CTGPPSAPRE | LWFEVQGSAL | MLHWRLPQEL | GGRGDLLFNV | VCKECGGHGE | PSSGGMCRRC |
| 430 | 440 | 450 | 460 | 470 | 480 |
| RDEVHFDPRQ | RGLTESRVLV | GGLRAHVPYI | LEVQAVNGVS | ELSPDPPQAA | AINVSTSHEV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| PSAVPVMHQV | SRAANSITVS | WPQPEQTNGN | ILDYQLRYYD | QAEDESHSFT | MTSETNTATV |
| 550 | 560 | 570 | 580 | 590 | 600 |
| TRLSPGHIYG | FQVRARTAAG | HGPYGGKVYF | QTLPQGELSS | QLPEKLSLVI | GSILGALAFL |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LLAAITVLAV | IFQRKRRGTG | YTEQLQQYSS | PGLGVKYYID | PSTYDDPCQA | IRELAREVDP |
| 670 | 680 | 690 | 700 | 710 | 720 |
| TYIKIEEVIG | AGSFGEVRRG | RLQPRGRREQ | AVAIQALWAG | GAESLKMTFL | GRAALLGQFQ |
| 730 | 740 | 750 | 760 | 770 | 780 |
| HPNILRLEGV | VTKSRPVMVL | TELMELGPLD | SFLRQREGQF | SSLQLVAMQR | GVAAAMQYLS |
| 790 | 800 | 810 | 820 | 830 | 840 |
| SFAFVHRALS | ARSVLVNSHL | VCKVARLGHS | PQGSSSLLRW | AAPEVITHGK | YTTSSDVWSF |
| 850 | 860 | 870 | 880 | 890 | 900 |
| GILMWEVMSY | GERPYWDMNE | QEVLNAIEQE | FRLPPPPGCP | PGLHLLMLDT | WQKDRARRPH |
| 910 | 920 | 930 | 940 | 950 | 960 |
| FDQLVAAFDK | MIRKPDTLQA | EGGSGDRPSQ | ALLNPVALDF | PCLDSPQAWL | SAIGLECYQD |
| 970 | 980 | 990 | 1000 | 1010 | |
| NFSKFGLSTF | SDVAQLSLED | LPGLGITLAG | HQKKLLHNIQ | LLQQHLRQPG | SVEV |