O07581
Gene name |
yhdL (BSU09510) |
Protein name |
Probable anti-sigma-M factor YhdL |
Names |
|
Species |
Bacillus subtilis (strain 168) |
KEGG Pathway |
bsu:BSU09510 |
EC number |
|
Protein Class |
|
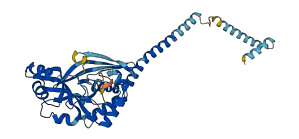
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for O07581
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O07581-F1 | Predicted | AlphaFoldDB |
No variants for O07581
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for O07581 | |||||
No associated diseases with O07581
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MMNEEFKKRF | DQYKNGEMSD | QEMTAFEEEL | EKLEVYQELI | DSELEDDNDW | DLSISPEKQK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| AILAYGKRKS | YLRISVLAVI | STLMILPLCT | LGSYLYYGMG | GKHSTGNEFM | ETAAVTVALT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| MPNVLVDTSG | LKSQVKLFGM | NTEFPLQKQI | GTKTAAVGNE | RVEMFYNKVK | APAVNYYDLE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VNKTGHYFTH | PSNKSEQTTA | KAEKTLSTLP | EGTVSEVYLS | YDRAYPTKDV | YNKFKGYDVS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FLWNAIETEK | NTNKTASTEP | LGYPGKDSKF | LAALNTKGKS | NGDQFINALK | FMSKHEKWAQ |
| 310 | 320 | 330 | 340 | 350 | |
| VISKRKDLNV | DNRLDYVEKN | GVNVYGSVVT | GPTKEIQRML | KNKSVKSANV | GEVELWNW |