O01798
Gene name |
spe-8 |
Protein name |
Spermatocyte protein spe-8 |
Names |
Defective spermatogenesis protein spe-8 |
Species |
Caenorhabditis elegans |
KEGG Pathway |
cel:CELE_F53G12.6 |
EC number |
2.7.10.2: Protein-tyrosine kinases |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
361-383 (Activation loop from InterPro)
Target domain |
217-485 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
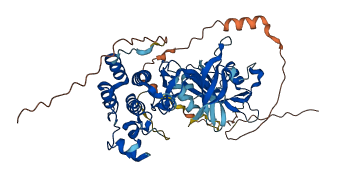
Autoinhibited structure

Activated structure

1 structures for O01798
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-O01798-F1 | Predicted | AlphaFoldDB |
No variants for O01798
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for O01798 | |||||
No associated diseases with O01798
7 regional properties for O01798
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 217 - 485 | IPR000719 |
| domain | SH2 domain | 112 - 205 | IPR000980 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 218 - 473 | IPR001245 |
| active_site | Tyrosine-protein kinase, active site | 340 - 352 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 223 - 250 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 217 - 475 | IPR020635 |
| domain | Fes/Fps/Fer, SH2 domain | 109 - 195 | IPR035849 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.10.2 | Protein-tyrosine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction: ATP + protein L-tyrosine = ADP + protein L-tyrosine phosphate by a non-membrane spanning protein. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| spermatid development | The process whose specific outcome is the progression of a spermatid over time, from its formation to the mature structure. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P16591 | FER | Tyrosine-protein kinase Fer | Homo sapiens (Human) | PR |
| P70451 | Fer | Tyrosine-protein kinase Fer | Mus musculus (Mouse) | PR |
| P09760 | Fer | Tyrosine-protein kinase Fer | Rattus norvegicus (Rat) | PR |
| Q8I7I5 | rol-3 | Protein roller-3 | Caenorhabditis elegans | PR |
| O45539 | src-2 | Tyrosine protein-kinase src-2 | Caenorhabditis elegans | SS |
| O16262 | nipi-4 | Protein nipi-4 | Caenorhabditis elegans | PR |
| P03949 | abl-1 | Tyrosine-protein kinase abl-1 | Caenorhabditis elegans | SS |
| G5ECJ6 | csk-1 | Tyrosine-protein kinase csk-1 | Caenorhabditis elegans | SS |
| G5EE56 | src-1 | Tyrosine protein-kinase src-1 | Caenorhabditis elegans | SS |
| Q95YD4 | kin-32 | Inactive tyrosine-protein kinase kin-32 | Caenorhabditis elegans | PR |
| Q2MHE4 | HT1 | Serine/threonine/tyrosine-protein kinase HT1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MRSKSSEGDL | QPEDTQSRED | KETTATYSED | TKPETQKERN | AALDNLAKTP | IQLVVQPTPL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TPAITPCEAP | PPPPPPKPSS | DNNNSKRLKV | KDQLIEVPSD | EVGRVENNID | NFPFYHGFMG |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RNECEAMLSN | HGDFLIRMTE | IGKRVAYVIS | IKWKYQNIHV | LVKRTKTKKL | YWTKKYAFKS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ICELIAYHKR | NHKPIYEGMT | LICGLARHGW | QLNNEQVTLN | KKLGEGQFGE | VHKGSLKTSV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| FAAPVTVAVK | TLHQNHLSAN | EKILFLREAN | VMLTLSHPNV | IKFYGVCTMK | EPIMIVMEFC |
| 310 | 320 | 330 | 340 | 350 | 360 |
| DGKSLEDALL | SKEEKVSAED | KILYLFHAAC | GIDYLHGKQV | IHRDIAARNC | LLNSKKILKI |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SDFGLSVKGV | AIKERKGGCL | PVKYMAPETL | KKGLYSTASD | IYSYGALMYE | VYTDGKTPFE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TCGLRGNELR | KAIIGKRISL | AVEVELPVFI | ANIFEQSRQY | ETEDRISSKQ | IIQIFKEEVG |
| 490 | 500 | 510 | |||
| FHEIETSGIL | HKLVNSLPRI | HNKERKPAAV | AV |