O00762
Gene name |
UBE2C (UBCH10) |
Protein name |
Ubiquitin-conjugating enzyme E2 C |
Names |
(E3-independent) E2 ubiquitin-conjugating enzyme C, E2 ubiquitin-conjugating enzyme C, UbcH10, Ubiquitin carrier protein C, Ubiquitin-protein ligase C |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:11065 |
EC number |
2.3.2.23: Aminoacyltransferases |
Protein Class |
|
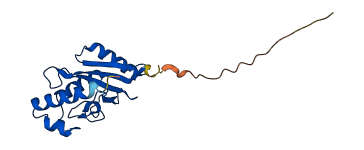
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

8 structures for O00762
148 variants for O00762
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
CA409185301 rs1225528823 |
3 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs984421119 CA315616073 |
5 | N>T | No |
ClinGen gnomAD |
|
|
rs561525053 CA315616091 |
6 | R>G | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9880537 rs561525053 |
6 | R>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs867231207 CA315616095 |
7 | D>A | No |
ClinGen Ensembl |
|
|
CA409185340 rs763421388 |
10 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763421388 CA9880539 |
10 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs763421388 CA315616099 |
10 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
| TCGA novel | 11 | T>missing | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA409185349 rs1243957339 |
11 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
rs1202446446 CA409185346 |
11 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1222721494 CA409185356 |
12 | S>R | No |
ClinGen TOPMed |
|
|
CA409185364 rs766882355 |
14 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766882355 CA409185365 |
14 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs766882355 CA9880540 |
14 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409185366 rs1186759513 |
14 | A>V | No |
ClinGen gnomAD |
|
|
rs1392036860 CA409185369 |
15 | A>T | No |
ClinGen gnomAD |
|
|
rs1271204292 CA409185377 |
16 | A>S | No |
ClinGen TOPMed |
|
|
rs1429651798 CA409185380 |
16 | A>V | No |
ClinGen gnomAD |
|
|
rs774646356 CA9880541 |
17 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs774646356 CA409185382 |
17 | R>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409185385 rs1376325872 |
17 | R>L | No |
ClinGen gnomAD |
|
|
rs1337052897 CA409185398 |
19 | G>E | No |
ClinGen gnomAD |
|
|
CA409185394 rs1467881661 |
19 | G>R | No |
ClinGen gnomAD |
|
|
CA409185401 rs1601038029 |
20 | A>T | No |
ClinGen Ensembl |
|
|
rs1387449323 CA409185414 |
22 | P>S | No |
ClinGen gnomAD |
|
|
CA9880543 rs11537645 |
23 | S>R | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA409185428 rs761413591 |
24 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs1368653521 CA409185430 |
24 | G>V | No |
ClinGen TOPMed |
|
|
rs761413591 CA9880545 |
24 | G>W | No |
ClinGen ExAC gnomAD |
|
| VAR_007694 | 25 | G>D | No | UniProt | |
|
CA409185438 rs1386174145 |
26 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs749925974 CA9880548 |
27 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409185444 rs749925974 |
27 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9880547 rs749925974 |
27 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1223889210 CA409185449 |
28 | R>W | No |
ClinGen gnomAD |
|
|
rs1267182417 CA409185467 |
31 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
rs1267182417 CA409185465 |
31 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
CA409185474 rs1356214083 |
32 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs1207003316 CA409185471 |
32 | G>S | No |
ClinGen gnomAD |
|
|
CA409185475 rs1356214083 |
32 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
rs761500077 CA9880565 |
36 | Q>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1190657829 CA409185517 |
37 | Q>* | No |
ClinGen gnomAD |
|
|
rs1258952908 CA409185523 |
38 | E>K | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed |
|
rs1207386492 CA409185532 |
39 | L>V | No |
ClinGen TOPMed |
|
|
CA409185543 rs1269104737 |
40 | M>I | No |
ClinGen TOPMed |
|
|
rs138058971 CA9880566 |
40 | M>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA409185549 rs1423586972 |
41 | T>I | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA409185547 rs1423586972 |
41 | T>N | No |
ClinGen gnomAD |
|
|
rs1223941923 CA409185558 |
43 | M>L | No |
ClinGen gnomAD |
|
|
rs545175408 CA9880567 |
43 | M>T | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA409185557 rs1223941923 |
43 | M>V | No |
ClinGen gnomAD |
|
|
CA9880588 rs766016081 |
47 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409185647 rs1189026288 |
54 | P>S | No |
ClinGen TOPMed |
|
|
CA315617320 rs377081477 |
56 | S>* | No |
ClinGen ESP TOPMed |
|
|
rs144831911 CA9880589 |
57 | D>N | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA9880590 rs759589512 |
59 | L>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409185688 rs1174298561 |
60 | F>S | No |
ClinGen gnomAD |
|
|
rs767562250 CA9880591 |
62 | W>C | No |
ClinGen ExAC gnomAD |
|
|
CA9880592 rs752601468 |
64 | G>A | No |
ClinGen ExAC gnomAD |
|
|
CA409185731 rs1225404499 |
66 | I>M | No |
ClinGen TOPMed |
|
|
CA409185733 rs1319994180 |
67 | H>D | No |
ClinGen gnomAD |
|
|
CA409185737 rs1433546288 |
67 | H>Q | No |
ClinGen gnomAD |
|
|
CA315617334 rs956674941 |
67 | H>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1319994180 CA409185734 |
67 | H>Y | No |
ClinGen gnomAD |
|
|
CA409185750 rs1271262675 |
69 | A>V | No |
ClinGen gnomAD |
|
|
rs779653538 CA9880649 |
73 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs746495033 CA9880650 |
74 | Y>* | No |
ClinGen ExAC gnomAD |
|
|
rs768695868 CA9880651 |
75 | E>D | No |
ClinGen ExAC gnomAD |
|
|
rs1448910805 CA409186132 |
77 | L>P | No |
ClinGen gnomAD |
|
|
CA9880652 rs547850953 |
78 | R>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA9880653 rs761667583 |
79 | Y>F | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9880654 rs769604009 |
80 | K>* | No |
ClinGen ExAC gnomAD |
|
|
rs772939191 CA9880655 |
80 | K>N | No |
ClinGen ExAC gnomAD |
|
|
rs763209065 CA9880656 |
82 | S>L | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA gnomAD |
|
CA9880658 rs751722028 |
84 | E>D | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs371616231 CA9880660 |
86 | P>H | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs759551578 CA9880659 |
86 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA9880661 rs753288094 |
87 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9880662 rs756640882 |
87 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA9880664 rs754226233 |
89 | Y>* | No |
ClinGen ExAC |
|
|
rs778333049 CA9880663 |
89 | Y>S | No |
ClinGen ExAC gnomAD |
|
|
rs374439898 CA315618202 |
90 | P>A | No |
ClinGen ESP |
|
|
rs758135325 CA9880665 |
92 | N>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs367706641 CA9880668 |
95 | T>A | No |
ClinGen ESP ExAC gnomAD |
|
|
rs367706641 CA409186238 |
95 | T>S | No |
ClinGen ESP ExAC gnomAD |
|
|
CA9880669 rs780709357 |
96 | V>M | No |
ClinGen ExAC gnomAD |
|
|
CA9880671 rs769693911 |
97 | K>N | No |
ClinGen ExAC gnomAD |
|
|
CA315618220 rs976266522 |
97 | K>R | No |
ClinGen TOPMed |
|
|
CA409186268 rs1198408107 |
100 | T>A | No |
ClinGen gnomAD |
|
|
CA9880672 COSM1027285 rs773029038 |
100 | T>M | Variant assessed as Somatic; 0.0 impact. endometrium [NCI-TCGA, Cosmic] | No |
ClinGen cosmic curated ExAC NCI-TCGA gnomAD |
|
CA409186278 rs1254022907 |
101 | P>L | No |
ClinGen TOPMed gnomAD |
|
|
rs932190109 CA315618263 |
103 | Y>F | No |
ClinGen TOPMed gnomAD |
|
|
rs770704295 CA409186312 |
106 | N>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9880676 rs770704295 |
106 | N>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
CA9880678 rs759764860 |
107 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1225682574 CA409186331 |
109 | T>N | No |
ClinGen gnomAD |
|
|
CA9880679 rs767520518 |
111 | G>D | No |
ClinGen ExAC gnomAD |
|
|
rs761307294 CA9880683 |
115 | L>P | No |
ClinGen ExAC gnomAD |
|
|
CA9880684 rs764691584 |
121 | K>T | No |
ClinGen ExAC gnomAD |
|
|
CA315618299 rs1033161095 |
122 | W>R | No |
ClinGen Ensembl |
|
|
rs1244942351 CA409186433 |
124 | A>S | No |
ClinGen TOPMed |
|
| TCGA novel | 124 | A>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9880686 COSM188841 rs757572822 |
125 | L>P | large_intestine Variant assessed as Somatic; impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated ExAC NCI-TCGA TOPMed gnomAD |
|
rs757572822 CA9880687 |
125 | L>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs148799246 CA9880690 |
126 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA9880689 rs148799246 |
126 | Y>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA315618344 rs7352110 |
129 | R>G | No |
ClinGen Ensembl |
|
|
rs371311367 CA409186469 |
130 | T>N | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs371311367 CA9880692 |
130 | T>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA315618347 rs201176918 |
131 | I>M | No |
ClinGen 1000Genomes |
|
|
rs767584548 CA315618348 |
133 | L>F | No |
ClinGen Ensembl |
|
|
rs777703076 CA9880693 |
136 | Q>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA315618357 rs902257654 |
138 | L>F | No |
ClinGen TOPMed |
|
|
rs1163401493 CA409186528 |
140 | G>E | No |
ClinGen TOPMed |
|
|
CA315618441 rs1036918950 |
141 | E>D | No |
ClinGen Ensembl |
|
|
rs1313636957 CA409186555 |
142 | P>R | No |
ClinGen gnomAD |
|
|
rs142378082 CA315618445 |
145 | D>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
rs1274157914 CA409186591 |
146 | S>N | No |
ClinGen gnomAD |
|
|
CA409186622 rs1272519354 |
150 | T>K | No |
ClinGen gnomAD |
|
|
CA9880719 rs201125748 |
154 | E>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1198011557 CA409186654 |
155 | L>I | No |
ClinGen TOPMed |
|
|
CA9880720 rs776802939 |
156 | W>R | No |
ClinGen ExAC gnomAD |
|
|
rs762275535 CA9880721 |
157 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs770303252 CA9880722 |
157 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA409186678 CA9880723 rs61760191 |
158 | N>K | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
| TCGA novel | 158 | N>T | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA9880724 rs763391914 |
159 | P>T | No |
ClinGen ExAC gnomAD |
|
|
rs373084608 CA9880725 |
160 | T>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA409186704 rs774676127 |
161 | A>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752378234 CA9880726 |
161 | A>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752378234 CA409186689 |
161 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA9880744 rs774676127 |
161 | A>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA409186734 rs1318325554 |
165 | Y>F | No |
ClinGen gnomAD |
|
|
CA409186743 rs753350058 |
167 | Q>* | No |
ClinGen ExAC gnomAD |
|
|
CA9880747 rs753350058 |
167 | Q>E | No |
ClinGen ExAC gnomAD |
|
|
rs1029457888 CA315619068 |
167 | Q>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen Ensembl NCI-TCGA |
|
CA9880748 rs761402285 |
168 | E>K | No |
ClinGen ExAC |
|
|
CA315619079 rs202149476 |
169 | T>A | No |
ClinGen 1000Genomes |
|
|
CA409186755 rs202149476 |
169 | T>P | No |
ClinGen 1000Genomes |
|
|
CA409186763 rs1568717224 |
170 | Y>S | No |
ClinGen Ensembl |
|
|
rs955166544 CA315619099 |
173 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
CA9880750 rs376878469 |
174 | V>G | No |
ClinGen ESP ExAC gnomAD |
|
|
CA9880751 rs758357409 |
175 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA409186809 rs1324101765 |
177 | Q>* | No |
ClinGen TOPMed gnomAD |
|
|
rs766325761 CA9880752 |
177 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
CA409186820 rs1282135234 |
178 | E>D | No |
ClinGen gnomAD |
|
|
rs985255663 CA315619134 |
178 | E>G | No |
ClinGen Ensembl |
No associated diseases with O00762
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.23 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
6 GO annotations of cellular component
| Name | Definition |
|---|---|
| anaphase-promoting complex | A ubiquitin ligase complex that degrades mitotic cyclins and anaphase inhibitory protein, thereby triggering sister chromatid separation and exit from mitosis. Substrate recognition by APC occurs through degradation signals, the most common of which is termed the Dbox degradation motif, originally discovered in cyclin B. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| ubiquitin ligase complex | A protein complex that includes a ubiquitin-protein ligase and enables ubiquitin protein ligase activity. The complex also contains other proteins that may confer substrate specificity on the complex. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ubiquitin conjugating enzyme activity | Isoenergetic transfer of ubiquitin from one protein to another via the reaction X-ubiquitin + Y -> Y-ubiquitin + X, where both the X-ubiquitin and Y-ubiquitin linkages are thioester bonds between the C-terminal glycine of ubiquitin and a sulfhydryl side group of a cysteine residue. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond: an isopeptide bond between the C-terminal glycine of ubiquitin and the epsilon-amino group of lysine residues in the substrate or, in the linear extension of ubiquitin chains, a peptide bond the between the C-terminal glycine and N-terminal methionine of ubiquitin residues. |
| ubiquitin-like protein ligase binding | Binding to a ubiquitin-like protein ligase, such as ubiquitin-ligase. |
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
12 GO annotations of biological process
| Name | Definition |
|---|---|
| anaphase-promoting complex-dependent catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, with ubiquitin-protein ligation catalyzed by the anaphase-promoting complex, and mediated by the proteasome. |
| cell division | The process resulting in division and partitioning of components of a cell to form more cells; may or may not be accompanied by the physical separation of a cell into distinct, individually membrane-bounded daughter cells. |
| exit from mitosis | The cell cycle transition where a cell leaves M phase and enters a new G1 phase. M phase is the part of the mitotic cell cycle during which mitosis and cytokinesis take place. |
| free ubiquitin chain polymerization | The process of creating free ubiquitin chains, compounds composed of a large number of ubiquitin monomers. These chains are not conjugated to a protein. |
| positive regulation of exit from mitosis | Any process that activates or increases the rate of progression from anaphase/telophase (high mitotic CDK activity) to G1 (low mitotic CDK activity). |
| positive regulation of ubiquitin protein ligase activity | Any process that activates or increases the frequency, rate or extent of ubiquitin protein ligase activity. |
| protein K11-linked ubiquitination | A protein ubiquitination process in which ubiquitin monomers are attached to a protein, and then ubiquitin polymers are formed by linkages between lysine residues at position 11 of the ubiquitin monomers. K11-linked polyubiquitination targets the substrate protein for degradation. The anaphase-promoting complex promotes the degradation of mitotic regulators by assembling K11-linked polyubiquitin chains. |
| protein K48-linked ubiquitination | A protein ubiquitination process in which a polymer of ubiquitin, formed by linkages between lysine residues at position 48 of the ubiquitin monomers, is added to a protein. K48-linked ubiquitination targets the substrate protein for degradation. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
| regulation of mitotic metaphase/anaphase transition | Any process that modulates the frequency, rate or extent of the cell cycle process in which a cell progresses from metaphase to anaphase during mitosis, triggered by the activation of the anaphase promoting complex by Cdc20/Sleepy homolog which results in the degradation of Securin. |
| ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of a ubiquitin group, or multiple ubiquitin groups, to the protein. |
12 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q32PA5 | UBE2C | Ubiquitin-conjugating enzyme E2 C | Bos taurus (Bovine) | PR |
| Q9H8T0 | AKTIP | AKT-interacting protein | Homo sapiens (Human) | PR |
| Q9Y385 | UBE2J1 | Ubiquitin-conjugating enzyme E2 J1 | Homo sapiens (Human) | PR |
| P61086 | UBE2K | Ubiquitin-conjugating enzyme E2 K | Homo sapiens (Human) | PR |
| Q9NPD8 | UBE2T | Ubiquitin-conjugating enzyme E2 T | Homo sapiens (Human) | PR |
| O14933 | UBE2L6 | Ubiquitin/ISG15-conjugating enzyme E2 L6 | Homo sapiens (Human) | PR |
| Q96LR5 | UBE2E2 | Ubiquitin-conjugating enzyme E2 E2 | Homo sapiens (Human) | PR |
| Q969T4 | UBE2E3 | Ubiquitin-conjugating enzyme E2 E3 | Homo sapiens (Human) | PR |
| Q7Z7E8 | UBE2Q1 | Ubiquitin-conjugating enzyme E2 Q1 | Homo sapiens (Human) | PR |
| A1L167 | UBE2QL1 | Ubiquitin-conjugating enzyme E2Q-like protein 1 | Homo sapiens (Human) | PR |
| Q16763 | UBE2S | Ubiquitin-conjugating enzyme E2 S | Homo sapiens (Human) | EV |
| Q9D1C1 | Ube2c | Ubiquitin-conjugating enzyme E2 C | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASQNRDPAA | TSVAAARKGA | EPSGGAARGP | VGKRLQQELM | TLMMSGDKGI | SAFPESDNLF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KWVGTIHGAA | GTVYEDLRYK | LSLEFPSGYP | YNAPTVKFLT | PCYHPNVDTQ | GNICLDILKE |
| 130 | 140 | 150 | 160 | 170 | |
| KWSALYDVRT | ILLSIQSLLG | EPNIDSPLNT | HAAELWKNPT | AFKKYLQETY | SKQVTSQEP |