O00571
Gene name |
DDX3X (DBX) |
Protein name |
ATP-dependent RNA helicase DDX3X |
Names |
CAP-Rf, DEAD box protein 3, X-chromosomal, DEAD box, X isoform, DBX, Helicase-like protein 2, HLP2 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:1654 |
EC number |
3.6.4.13: Acting on ATP; involved in cellular and subcellular movement |
Protein Class |
ATP-DEPENDENT RNA HELICASE DBP3 (PTHR47958) |
Descriptions
DEAD-box proteins utilize ATP to bind and remodel RNA and RNA-protein complexes. The Ded1/DDX3 subfamily of DEAD-box proteins is of particular interest as their function during protein translation, are essential for viability, and are frequently altered in human malignancies.
There is a unique interdomain interaction between the two ATPase domains (DEAD box helicase domain and helicase domain) in which the C-terminal helicase domain clashes with the RNA-binding surface. Destabilizing this interaction accelerates RNA duplex unwinding, suggesting that it is present in solution and inhibitory for catalysis. The N-terminal extension to the DEAD box helicase interacts with DEAD box helicase domain and C-terminal helicase domain, and stabilizes the autoinhibited state of DDX3X.
Autoinhibitory domains (AIDs)
Target domain |
204-391 (DEAD box helicase domain) |
Relief mechanism |
|
Assay |
Structural analysis |
Target domain |
414-575 (C-terminal helicase domain) |
Relief mechanism |
|
Assay |
Structural analysis |
Target domain |
204-391 (DEAD box helicase domain); 414-575 (C-terminal helicase domain) |
Relief mechanism |
|
Assay |
Structural analysis, Mutagenesis experiment |
Accessory elements
No accessory elements
References
- Floor SN et al. (2016) "Autoinhibitory Interdomain Interactions and Subfamily-specific Extensions Redefine the Catalytic Core of the Human DEAD-box Protein DDX3", The Journal of biological chemistry, 291, 2412-21
- Zhang ZM et al. (2013) "Crystal structure of Prp5p reveals interdomain interactions that impact spliceosome assembly", Cell reports, 5, 1269-78
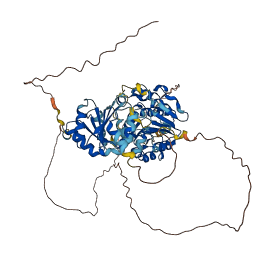
Autoinhibited structure
Activated structure
15 structures for O00571
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 2I4I | X-ray | 220 A | A | 168-582 | PDB |
| 2JGN | X-ray | 191 A | A/B/C | 409-580 | PDB |
| 3JRV | X-ray | 160 A | C/D/E | 71-90 | PDB |
| 4O2C | X-ray | 180 A | C | 2-10 | PDB |
| 4O2E | X-ray | 198 A | C/F | 2-10 | PDB |
| 4O2F | X-ray | 190 A | C/F | 3-10 | PDB |
| 4PX9 | X-ray | 231 A | A/B/C | 135-407 | PDB |
| 4PXA | X-ray | 320 A | A | 135-582 | PDB |
| 5E7I | X-ray | 222 A | A/B/C | 133-584 | PDB |
| 5E7J | X-ray | 223 A | A | 133-584 | PDB |
| 5E7M | X-ray | 230 A | A | 133-584 | PDB |
| 6CZ5 | X-ray | 300 A | A | 132-607 | PDB |
| 6O5F | X-ray | 250 A | A/B | 132-607 | PDB |
| 7LIU | X-ray | 300 A | A/B | 135-582 | PDB |
| AF-O00571-F1 | Predicted | AlphaFoldDB |
300 variants for O00571
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
RCV000622971 rs1555950665 |
1 | M>I | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1555950676 RCV000598857 RCV001260599 CA412761583 |
14 | Q>* | Intellectual disability [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1602119305 RCV000990792 |
35 | K>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412762885 RCV000656409 rs1555951993 |
38 | Y>C | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1341824034 RCV001253093 |
41 | P>H | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1064793796 RCV000483467 CA16621378 RCV001253449 |
41 | P>S | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1602122237 RCV000850466 |
51 | G>missing | Marfanoid habitus and intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412764733 RCV000990794 rs1602126980 |
58 | S>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000416474 rs1057519431 |
65 | D>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs886041589 CA10603727 RCV001266460 RCV000371670 |
78 | S>* | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001093533 rs1064795656 RCV000478243 CA16621380 |
79 | R>K | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs1131691299 RCV000493746 RCV000678322 |
85 | F>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1602127262 RCV000990795 |
90 | S>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000416457 rs1057519446 CA16044242 |
121 | R>L | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar TOPMed dbSNP gnomAD |
|
rs2063866727 RCV001267612 |
127 | W>* | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001266038 rs2063868825 |
176 | P>R | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001336293 rs2063875155 |
182 | F>V | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000623644 rs1555953152 CA412767815 |
190 | I>S | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs875989803 RCV000211113 CA10576137 |
193 | G>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA412768034 RCV000623907 rs1555953166 |
206 | V>M | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000209890 CA353430 rs869312692 |
207 | Q>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000493357 rs1131691571 RCV001249289 |
214 | I>missing | DDX3X-Related Disorder [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063876114 RCV001093537 |
214 | I>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063876197 RCV001197879 |
215 | K>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063876393 RCV001252462 |
219 | D>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001252463 rs2063876594 |
225 | Q>R | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063884615 RCV001253654 |
231 | T>S | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs796052223 VAR_075732 CA319624 |
233 | A>V | MRXSSB [UniProt] | Yes |
ClinGen UniProt Ensembl dbSNP |
|
rs796052224 CA319626 VAR_075734 |
235 | L>P | MRXSSB [UniProt] | Yes |
ClinGen UniProt Ensembl dbSNP |
|
rs752738546 CA211955 RCV000209908 |
249 | E>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
rs1555953398 RCV000505230 |
257 | N>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063889019 RCV001254065 |
260 | Y>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412770023 RCV000623442 rs1555953406 |
262 | R>C | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA412770153 rs1267519974 RCV000677413 |
274 | P>S | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar dbSNP gnomAD |
|
rs2063889558 RCV001336294 |
275 | T>M | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1602131859 RCV001093538 |
276 | R>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1602131859 RCV001836903 RCV000824883 |
277 | E>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1602131872 RCV000824881 |
278 | L>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001199268 rs2063889955 |
286 | A>D | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs1064796820 CA16621383 RCV000479176 RCV001824805 |
288 | K>E | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000190553 CA358846 rs869320681 |
291 | Y>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000718415 RCV000519583 CA412770619 rs1555953488 |
292 | R>* | History of neurodevelopmental disorder [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA412770694 rs1602132216 RCV000824882 |
296 | R>P | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000211100 rs875989802 CA10576138 |
302 | G>S | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1453153749 RCV001260602 RCV001332990 RCV001310708 |
311 | R>* | Intellectual disability Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001706658 rs1555953527 RCV000523985 RCV001267128 |
321 | V>missing | Intellectual disability, X-linked 102 Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001260597 rs2063893070 |
325 | G>missing | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001007875 rs1555953548 RCV000624280 CA412771087 |
326 | R>C | Intellectual disability, X-linked 102 Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs797045025 RCV000190552 CA204535 RCV000521776 RCV000623237 VAR_075736 |
326 | R>H | MRXSSB; loss-of-function mutation affecting regulation of Wnt signaling Intellectual disability, X-linked 102 Inborn genetic diseases [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000505622 rs1555953796 CA412771237 |
345 | V>L | Medulloblastoma [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1057518707 CA16043708 VAR_075737 RCV000414932 |
351 | R>Q | Intellectual disability, X-linked 102 MRXSSB [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000190554 CA204537 rs797045026 VAR_075738 |
362 | R>C | Intellectual disability, X-linked 102 MRXSSB [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000505208 rs1555953819 RCV001420280 |
367 | Q>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA204531 rs796052231 RCV000190550 VAR_075739 |
376 | R>C | MRXSSB; also found as a somatic mutation in medulloblastoma; loss of ATPase activity; increased interaction with CSNK1E in the absence of dsRNA; contrary to wild-type protein, strongly interacts with CSNK1A1 and CSNK1D in vivo; strongly increased ability to activate CSNK1E kinase activity, leading to increased DVL phosphorylation, thereby activating Wnt/beta-catenin signaling; increased RNA-binding; no effect on subcellular location Intellectual disability, X-linked 102 [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001093534 rs2063908570 |
376 | R>H | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000995528 rs1602134248 CA412771509 |
383 | A>G | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001706923 VAR_075740 CA319636 rs796052232 |
392 | L>P | Intellectual disability, X-linked 102 MRXSSB [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs1602134468 RCV000850545 |
394 | R>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001267423 rs2063910538 |
397 | L>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001260603 rs2063910640 |
400 | Y>* | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000624401 RCV000433202 CA16608470 rs1057523822 |
409 | G>D | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs796052233 CA319638 VAR_075741 |
417 | Q>P | MRXSSB [UniProt] | Yes |
ClinGen UniProt Ensembl dbSNP |
|
rs1555953882 RCV000624480 |
426 | D>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001266926 rs2063911610 |
434 | L>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063911698 RCV001269284 |
435 | L>P | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001260596 rs2063922414 |
448 | V>E | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001260600 rs2063922977 |
467 | A>missing | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001265801 RCV001751532 rs2063923221 |
472 | H>P | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000486655 RCV001266615 VAR_075742 CA16621388 rs1064794574 |
475 | R>G | Inborn genetic diseases MRXSSB [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV001267145 rs1555954105 |
477 | Q>* | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412775358 RCV000677395 rs1555954105 |
477 | Q>E | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs2063923605 RCV001093536 |
479 | D>VS | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001257983 CA412775457 rs1569240005 RCV000779659 |
480 | R>T | Congenital cerebellar hypoplasia Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs2063923749 RCV001252461 |
483 | A>D | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063923789 RCV001253728 |
485 | H>P | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA319640 rs796052234 RCV001266548 RCV000505799 |
488 | R>C | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000190112 RCV000656416 rs796052235 VAR_075744 CA319642 |
488 | R>H | Intellectual disability, X-linked 102 MRXSSB [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000660645 rs1555954154 CA412775876 |
496 | V>M | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs796052236 RCV001857666 CA319644 RCV001265760 |
497 | A>V | Inborn genetic diseases [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001093535 rs2063927503 |
504 | G>E | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs797045024 RCV000190551 CA204533 VAR_075745 |
507 | I>T | MRXSSB; loss-of-function mutation affecting regulation of Wnt signaling Intellectual disability, X-linked 102 [UniProt, ClinVar] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000190107 RCV000718596 rs796052230 RCV001256979 RCV001093539 |
512 | H>missing | Rare genetic intellectual disability Intellectual disability, X-linked 102 History of neurodevelopmental disorder [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063927827 RCV001260758 |
513 | V>missing | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
CA412776373 rs1569240261 RCV000717944 |
513 | V>A | History of neurodevelopmental disorder [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001260601 rs2063927856 |
513 | V>I | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063927856 RCV001310221 |
513 | V>L | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
CA319630 rs796052226 RCV000190103 VAR_075747 RCV001264651 |
514 | I>T | MRXSSB [UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
rs2063928003 RCV001260759 |
515 | N>D | Intellectual disability [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000824983 rs1602136369 |
522 | I>missing | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs766409654 RCV001196847 |
525 | Y>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001824802 CA16621392 rs1064795323 RCV000484659 |
528 | R>C | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000505686 rs1555954272 CA412776884 |
530 | G>C | Medulloblastoma [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
CA412776919 rs1555954275 RCV000505558 |
531 | R>H | Medulloblastoma [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1555954284 RCV000625959 CA412776975 |
534 | R>C | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1555954284 CA412776972 RCV000856741 |
534 | R>G | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV000624510 rs1555954287 |
535 | V>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000482989 rs1064794993 CA16621394 RCV001796072 |
559 | L>H | DDX3X-related X-linked intellectual disability [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs1555954380 RCV001255961 RCV001260756 RCV000599287 |
560 | L>missing | Intellectual disability Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
VAR_075750 RCV000622529 RCV000416455 RCV001093496 rs1057519430 CA16044243 |
568 | P>L | Intellectual disability, X-linked 102 Inborn genetic diseases MRXSSB [ClinVar, UniProt] | Yes |
ClinGen ClinVar UniProt Ensembl dbSNP |
|
RCV000509440 rs1555954388 CA412777821 |
570 | W>* | DDX3X-Related Disorder [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
RCV001281103 rs2063933550 |
576 | Y>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinVar dbSNP |
|
rs2063933817 RCV001089641 |
583 | S>missing | Global developmental delay [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV000767345 RCV000328364 rs886041705 CA10603593 |
603 | R>* | Intellectual disability, X-linked 102 [ClinVar] | Yes |
ClinGen ClinVar Ensembl dbSNP |
|
rs2063940433 RCV001267361 |
613 | F>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
RCV001266343 rs2063941013 |
620 | S>missing | Inborn genetic diseases [ClinVar] | Yes |
ClinVar dbSNP |
|
CA10389281 rs776297102 |
5 | A>S | No |
ClinGen ExAC gnomAD |
|
|
rs1158832456 COSM1132302 CA412762246 |
17 | A>G | prostate [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
CA412762287 rs1569233520 |
20 | D>Y | No |
ClinGen Ensembl |
|
|
CA412762358 rs1309663175 |
22 | N>S | No |
ClinGen TOPMed |
|
|
RCV001171565 rs2063786541 |
25 | D>missing | No |
ClinVar dbSNP |
|
|
rs1175361408 CA412762448 |
25 | D>E | No |
ClinGen gnomAD |
|
|
RCV001268169 rs2063787069 |
26 | N>missing | No |
ClinVar dbSNP |
|
|
CA412762492 rs1602119250 RCV000999402 |
27 | Q>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1602119274 RCV001009107 |
28 | S>missing | No |
ClinVar dbSNP |
|
|
RCV000368239 rs886041769 |
28 | S>missing | No |
ClinVar dbSNP |
|
|
rs2063787500 RCV001093492 |
32 | T>missing | No |
ClinVar dbSNP |
|
|
rs752354649 CA10389328 |
34 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1555951992 RCV000626446 |
38 | Y>missing | No |
ClinVar dbSNP |
|
|
RCV000478349 CA16621377 rs1064793858 |
40 | P>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412762929 RCV000999403 rs1602122138 |
40 | P>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1341824034 CA412762953 |
41 | P>L | No |
ClinGen gnomAD |
|
|
RCV000760483 CA412763072 rs1569234653 |
46 | R>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs774308886 CA10389354 |
49 | T>I | No |
ClinGen ExAC gnomAD |
|
|
CA16621379 RCV000484981 rs1064796382 |
50 | K>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412764630 rs1263310293 |
51 | G>D | No |
ClinGen gnomAD |
|
|
CA412764671 rs1187201543 |
54 | D>N | No |
ClinGen gnomAD |
|
|
rs1602126956 RCV001008615 |
56 | D>missing | No |
ClinVar dbSNP |
|
|
CA329018673 rs1012287078 |
56 | D>G | No |
ClinGen TOPMed |
|
|
CA412764701 rs1198871978 |
56 | D>N | No |
ClinGen gnomAD |
|
|
CA10389375 rs749047477 |
57 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs1555952639 CA412764730 RCV000497901 |
58 | S>A | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA10389377 rs778806111 |
61 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA412764771 rs1369281442 |
61 | S>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1453386913 CA412764842 |
66 | K>R | No |
ClinGen gnomAD |
|
|
rs2063850491 RCV001200319 |
67 | D>missing | No |
ClinVar dbSNP |
|
|
rs1250603833 CA412764875 |
68 | A>V | No |
ClinGen TOPMed |
|
|
rs1555952659 RCV000514961 |
69 | Y>missing | No |
ClinVar dbSNP |
|
|
rs772081578 CA10389379 |
71 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA412764957 rs1289923555 |
75 | R>C | No |
ClinGen TOPMed |
|
|
CA412764961 rs1569236301 |
75 | R>H | No |
ClinGen Ensembl |
|
|
rs1312827755 CA412764970 |
76 | S>G | No |
ClinGen gnomAD |
|
|
CA10389381 rs774816127 |
78 | S>P | No |
ClinGen ExAC gnomAD |
|
|
CA658799707 RCV000598952 rs1555952685 |
81 | K>T* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412765097 rs1236484806 |
83 | S>G | No |
ClinGen gnomAD |
|
|
CA329018703 rs892274119 |
84 | F>C | No |
ClinGen TOPMed gnomAD |
|
|
rs763397133 CA10389385 |
88 | R>H | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA412765242 rs1602127249 |
89 | G>E | No |
ClinGen Ensembl |
|
|
rs766794672 CA10389386 |
90 | S>G | No |
ClinGen ExAC gnomAD |
|
|
rs1374495935 CA412765330 |
94 | G>E | No |
ClinGen TOPMed |
|
|
rs1479548397 CA412765588 |
99 | R>C | No |
ClinGen gnomAD |
|
|
CA329019240 rs11552983 |
100 | G>V | No |
ClinGen Ensembl |
|
|
CA412765623 rs1311783625 |
101 | R>Q | No |
ClinGen TOPMed |
|
|
rs1175660869 CA412765621 |
101 | R>W | No |
ClinGen TOPMed gnomAD |
|
|
CA412765677 rs1602129054 |
104 | Y>D | No |
ClinGen Ensembl |
|
|
CA412765739 rs1301463504 |
107 | I>V | No |
ClinGen TOPMed |
|
|
rs1418745394 CA412765774 |
109 | S>R | No |
ClinGen gnomAD |
|
|
CA412765806 rs1444942370 |
110 | R>C | No |
ClinGen TOPMed |
|
|
CA329019261 rs1047424997 |
112 | D>H | No |
ClinGen Ensembl |
|
|
rs1296063643 CA412765922 |
115 | G>D | No |
ClinGen gnomAD |
|
|
CA10389404 rs747737720 |
115 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs774528109 CA10389406 |
117 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs759994098 CA10389407 |
118 | K>R | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10389409 rs776128256 |
121 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1057519446 CA412766080 |
121 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1057519446 CA412766076 |
121 | R>P | No |
ClinGen TOPMed gnomAD |
|
|
rs776128256 CA412766064 |
121 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1057518171 RCV000413210 |
124 | N>missing | No |
ClinVar dbSNP |
|
|
RCV001009206 rs1602129221 |
125 | S>missing | No |
ClinVar dbSNP |
|
|
CA10389410 rs760654985 |
127 | W>C | No |
ClinGen ExAC gnomAD |
|
|
rs2063866799 RCV001093493 |
130 | K>missing | No |
ClinVar dbSNP |
|
|
rs1439789058 CA412766306 |
130 | K>T | No |
ClinGen gnomAD |
|
|
rs758942832 CA329019310 |
139 | P>L | No |
ClinGen Ensembl |
|
|
RCV001268469 rs2063867157 |
148 | Q>* | No |
ClinVar dbSNP |
|
|
CA16621381 rs1064794404 RCV000484338 |
148 | Q>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs769546741 RCV000627323 CA412767141 |
163 | Y>* | No |
ClinGen ClinVar ExAC TOPMed dbSNP gnomAD |
|
|
rs777522116 CA10389425 |
165 | D>G | No |
ClinGen ExAC gnomAD |
|
|
CA412767270 rs1395981267 |
170 | A>T | No |
ClinGen gnomAD |
|
|
rs898635517 CA329019422 |
171 | T>I | No |
ClinGen Ensembl |
|
|
CA329019442 rs997047370 |
178 | H>R | No |
ClinGen TOPMed |
|
|
CA10389427 rs772541227 |
181 | S>T | No |
ClinGen ExAC |
|
|
RCV001093494 rs2063875302 |
194 | N>S | No |
ClinVar dbSNP |
|
|
CA10389443 rs375996245 |
196 | E>K | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
RCV000522090 CA412767937 rs1555953158 |
198 | T>P | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs755750366 CA10389444 |
199 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs777274761 RCV001269948 |
200 | Y>* | No |
ClinVar dbSNP |
|
|
rs796052237 CA319646 |
200 | Y>* | No |
ClinGen Ensembl |
|
|
rs1555953169 RCV000498348 |
208 | K>missing | No |
ClinVar dbSNP |
|
|
RCV001171597 rs2063875918 |
208 | K>missing | No |
ClinVar dbSNP |
|
|
rs886039719 CA10588780 RCV000255056 |
212 | P>H | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412768161 rs1298731437 |
215 | K>R | No |
ClinGen TOPMed |
|
|
CA412768176 rs1283619077 |
216 | E>G | No |
ClinGen gnomAD |
|
|
rs1064796429 RCV000482407 CA16621382 |
220 | L>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs759442213 CA10389464 |
241 | Q>K | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1311554406 CA412769545 |
241 | Q>R | No |
ClinGen TOPMed |
|
|
rs1275844963 CA412769597 |
244 | S>L | No |
ClinGen TOPMed |
|
|
rs752738546 CA10389466 |
249 | E>K | No |
ClinGen ExAC gnomAD |
|
|
rs1131691455 RCV000492974 CA412769704 |
250 | A>T | No |
ClinGen ClinVar TOPMed dbSNP |
|
|
rs1555953333 RCV000656239 |
251 | L>missing | No |
ClinVar dbSNP |
|
|
rs1602131321 RCV001009290 |
251 | L>missing | No |
ClinVar dbSNP |
|
|
rs1334191780 CA412769738 |
251 | L>S | No |
ClinGen gnomAD |
|
|
rs756881734 CA10389470 |
254 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA412769803 rs1225804340 |
254 | M>R | No |
ClinGen TOPMed |
|
|
CA10389469 rs753317345 |
254 | M>V | No |
ClinGen ExAC gnomAD |
|
|
rs760617639 CA10389487 |
258 | G>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10389488 rs763958253 |
260 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA412770029 rs1218821680 |
262 | R>H | No |
ClinGen gnomAD |
|
|
CA329020223 rs1021585207 |
263 | R>C | No |
ClinGen TOPMed |
|
|
CA329020226 rs867967504 |
263 | R>H | No |
ClinGen Ensembl |
|
|
rs761300398 CA10389490 |
268 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
RCV000598898 rs1555953420 |
274 | P>missing | No |
ClinVar dbSNP |
|
|
RCV000762624 rs1569238251 |
275 | T>missing | No |
ClinVar dbSNP |
|
|
CA10389491 rs752332279 |
280 | V>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1425452704 CA412770437 |
287 | R>K | No |
ClinGen gnomAD |
|
|
rs1555953482 CA412770585 RCV000598714 |
290 | S>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA16621384 RCV000484846 rs1064796759 |
292 | R>Q | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1453153749 CA412770946 |
311 | R>G | No |
ClinGen gnomAD |
|
|
rs762545072 CA10389513 |
313 | L>M | No |
ClinGen ExAC gnomAD |
|
|
CA319628 rs796052225 |
315 | R>L | No |
ClinGen Ensembl |
|
|
rs756500897 CA10389515 |
318 | H>L | No |
ClinGen ExAC gnomAD |
|
|
rs778328870 CA10389516 |
319 | L>F | No |
ClinGen ExAC |
|
|
rs754240311 CA10389517 |
321 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA412771069 rs1237306159 |
323 | T>A | No |
ClinGen gnomAD |
|
|
CA16608468 RCV000434513 rs1057521603 |
323 | T>I | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412771091 RCV000523594 rs1555953550 |
327 | L>V | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1354285733 CA412771124 |
331 | M>I | No |
ClinGen gnomAD |
|
|
rs994574415 CA329020399 |
339 | D>N | No |
ClinGen Ensembl |
|
|
rs749156638 CA10389582 |
365 | V>A | No |
ClinGen ExAC |
|
|
CA412771410 rs1307578363 |
369 | T>S | No |
ClinGen TOPMed |
|
|
rs1602134217 CA412771444 |
374 | G>D | No |
ClinGen Ensembl |
|
|
rs1064794068 RCV000480302 CA16621385 |
382 | S>R | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412771531 rs1440305030 |
387 | K>E | No |
ClinGen TOPMed |
|
|
CA412771599 rs1354602308 |
391 | M>I | No |
ClinGen gnomAD |
|
|
CA10389602 rs753655574 |
394 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA412771629 RCV000519312 rs1555953865 |
394 | R>H | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1064794133 RCV000480376 |
402 | F>missing | No |
ClinVar dbSNP |
|
|
RCV000505734 rs796052238 |
404 | A>missing | No |
ClinVar dbSNP |
|
|
rs1064796364 RCV000477995 |
419 | V>missing | No |
ClinVar dbSNP |
|
|
CA10389609 rs768445879 |
427 | K>R | No |
ClinGen ExAC gnomAD |
|
|
rs1555953894 RCV000657547 |
429 | S>missing | No |
ClinVar dbSNP |
|
|
rs773694310 CA10389610 |
430 | F>Y | No |
ClinGen ExAC gnomAD |
|
|
RCV000190104 rs796052227 |
441 | D>missing | No |
ClinVar dbSNP |
|
|
CA412774062 rs1458715097 |
441 | D>G | No |
ClinGen gnomAD |
|
|
rs1555954081 CA412774176 RCV000497906 |
445 | L>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412774825 RCV001007997 rs1602135779 |
464 | E>* | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA10389630 rs746566984 |
474 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs1064794574 CA412775294 RCV000523821 |
475 | R>C | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
RCV000999404 rs1602135832 CA412775297 |
475 | R>H | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs939577582 CA329021453 |
478 | R>K | No |
ClinGen TOPMed gnomAD |
|
|
CA16621389 rs1064796827 RCV000478814 |
480 | R>G | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1555954122 RCV000627502 |
485 | H>missing | No |
ClinVar dbSNP |
|
|
CA412775626 rs1332205043 |
485 | H>Q | No |
ClinGen gnomAD |
|
|
RCV000327317 rs886041963 |
488 | R>missing | No |
ClinVar dbSNP |
|
|
CA10389636 rs774580948 |
491 | K>E | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10389635 rs774580948 |
491 | K>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA16621390 rs1064796924 RCV000485793 |
494 | I>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
RCV000413345 CA16043317 rs1057518255 |
495 | L>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA412775902 rs1602135997 |
497 | A>P | No |
ClinGen Ensembl |
|
|
CA412775929 rs200427211 RCV000489888 |
498 | T>I | No |
ClinGen ClinVar ExAC dbSNP gnomAD |
|
|
rs200427211 CA10389637 |
498 | T>K | No |
ClinGen ExAC gnomAD |
|
|
CA10389639 rs760784261 |
499 | A>E | No |
ClinGen ExAC gnomAD |
|
|
CA10389638 rs775524960 |
499 | A>T | No |
ClinGen ExAC gnomAD |
|
|
RCV001008488 rs1602136301 |
502 | A>missing | No |
ClinVar dbSNP |
|
|
CA16621391 rs1017583398 RCV000478952 |
505 | L>V | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1311231381 CA412776540 |
515 | N>S | No |
ClinGen TOPMed |
|
|
rs966045593 CA329021569 |
522 | I>V | No |
ClinGen Ensembl |
|
|
RCV000484531 rs1064795387 CA16621393 |
532 | T>M | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs2063928899 RCV001264625 |
534 | R>missing | No |
ClinVar dbSNP |
|
|
rs1232247926 CA412777412 |
550 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
RCV001269944 rs2063932925 |
553 | T>missing | No |
ClinVar dbSNP |
|
|
RCV000424111 rs1057521175 CA16608473 |
556 | L>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
CA10389684 rs759495182 |
559 | L>V | No |
ClinGen ExAC gnomAD |
|
|
rs767400037 CA10389685 |
561 | V>L | No |
ClinGen ExAC gnomAD |
|
|
RCV000659154 rs1555954386 CA412777767 |
567 | V>L | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs886043639 CA10605769 RCV000263868 |
571 | L>S | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs2063933502 RCV001093497 |
574 | M>V | No |
ClinVar dbSNP |
|
|
rs1602137066 CA412778055 |
580 | Y>H | No |
ClinGen Ensembl |
|
|
rs753372536 CA10389689 |
583 | S>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA329021853 rs887951133 |
585 | R>C | No |
ClinGen Ensembl |
|
|
CA412778254 rs1344106721 |
585 | R>H | No |
ClinGen TOPMed |
|
|
rs1175768430 CA412778325 |
587 | R>H | No |
ClinGen TOPMed gnomAD |
|
|
rs1463769342 CA412778543 |
591 | S>N | No |
ClinGen gnomAD |
|
|
rs761434235 CA10389713 |
594 | S>N | No |
ClinGen ExAC gnomAD |
|
|
rs751917713 CA10389715 |
597 | F>V | No |
ClinGen ExAC gnomAD |
|
|
rs755359362 CA10389716 |
607 | G>S | No |
ClinGen ExAC gnomAD |
|
|
rs781633738 CA10389717 |
608 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs769390253 CA10389718 |
609 | S>C | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs868308965 CA329022019 |
612 | S>R | No |
ClinGen Ensembl |
|
|
rs756678720 CA10389719 |
614 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA10389721 rs756197314 |
618 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA10389720 rs756197314 |
618 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs901961447 CA329022047 |
622 | R>H | No |
ClinGen gnomAD |
|
|
rs1555954556 CA412779418 RCV000523261 |
623 | S>I | No |
ClinGen ClinVar Ensembl dbSNP |
|
|
rs1264184971 CA412779430 |
624 | G>S | No |
ClinGen gnomAD |
|
|
rs1200557241 CA412779454 |
625 | G>R | No |
ClinGen gnomAD |
|
|
rs1476609971 CA412779584 |
629 | G>S | No |
ClinGen gnomAD |
|
|
CA10389724 rs745644072 |
629 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs1351543785 CA412779794 |
636 | G>R | No |
ClinGen gnomAD |
|
|
rs1385128223 CA412780720 |
639 | Y>C | No |
ClinGen TOPMed gnomAD |
|
|
CA329022125 rs1001358042 |
644 | N>S | No |
ClinGen gnomAD |
|
|
CA329022128 rs201813358 |
645 | S>I | No |
ClinGen Ensembl |
1 associated diseases with O00571
[MIM: 300958]: Intellectual developmental disorder, X-linked, syndromic, Snijders Blok type (MRXSSB)
A disorder characterized by mild to severe intellectual disability, hypotonia, movement disorders, behavior problems, corpus callosum hypoplasia, and epilepsy. Additionally, patients manifest variable non-neurologic features such as joint hyperlaxity, skin pigmentary abnormalities, cleft lip and/or palate, hearing and visual impairment, and precocious puberty. {ECO:0000269|PubMed:26235985}. Note=The disease is caused by variants affecting the gene represented in this entry.
Without disease ID
- A disorder characterized by mild to severe intellectual disability, hypotonia, movement disorders, behavior problems, corpus callosum hypoplasia, and epilepsy. Additionally, patients manifest variable non-neurologic features such as joint hyperlaxity, skin pigmentary abnormalities, cleft lip and/or palate, hearing and visual impairment, and precocious puberty. {ECO:0000269|PubMed:26235985}. Note=The disease is caused by variants affecting the gene represented in this entry.
5 regional properties for O00571
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| conserved_site | ATP-dependent RNA helicase DEAD-box, conserved site | 345 - 353 | IPR000629 |
| domain | Helicase, C-terminal | 414 - 575 | IPR001650 |
| domain | DEAD/DEAH box helicase domain | 204 - 391 | IPR011545 |
| domain | Helicase superfamily 1/2, ATP-binding domain | 199 - 418 | IPR014001 |
| domain | RNA helicase, DEAD-box type, Q motif | 180 - 208 | IPR014014 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.6.4.13 | Acting on ATP; involved in cellular and subcellular movement |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47958 | ATP-DEPENDENT RNA HELICASE DBP3 |
| PANTHER Subfamily | PTHR47958:SF188 | ATP-DEPENDENT RNA HELICASE DDX3X |
| PANTHER Protein Class | RNA helicase | |
| PANTHER Pathway Category | No pathway information available | |
15 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell leading edge | The area of a motile cell closest to the direction of movement. |
| centrosome | A structure comprised of a core structure (in most organisms, a pair of centrioles) and peripheral material from which a microtubule-based structure, such as a spindle apparatus, is organized. Centrosomes occur close to the nucleus during interphase in many eukaryotic cells, though in animal cells it changes continually during the cell-division cycle. |
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytoplasmic stress granule | A dense aggregation in the cytosol composed of proteins and RNAs that appear when the cell is under stress. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| ficolin-1-rich granule lumen | Any membrane-enclosed lumen that is part of a ficolin-1-rich granule. |
| lamellipodium | A thin sheetlike process extended by the leading edge of a migrating cell or extending cell process; contains a dense meshwork of actin filaments. |
| NLRP3 inflammasome complex | An inflammasome complex that consists of three components, NLRP3 (NALP3), PYCARD and caspase-1. It is activated upon exposure to whole pathogens, as well as a number of structurally diverse pathogen- and danger-associated molecular patterns (PAMPs and DAMPs) and environmental irritants. Whole pathogens demonstrated to activate the NLRP3 inflammasome complex include the fungi Candida albicans and Saccharomyces cerevisiae, bacteria that produce pore-forming toxins, including Listeria monocytogenes and Staphylococcus aureus, and viruses such as Sendai virus, adenovirus, and influenza virus. |
| nucleoplasm | That part of the nuclear content other than the chromosomes or the nucleolus. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| P granule | A small cytoplasmic, non-membranous RNA/protein complex aggregate in the primordial germ cells of many higher eukaryotes. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
| secretory granule lumen | The volume enclosed by the membrane of a secretory granule. |
21 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATP hydrolysis activity | Catalysis of the reaction: ATP + H2O = ADP + H+ phosphate. ATP hydrolysis is used in some reactions as an energy source, for example to catalyze a reaction or drive transport against a concentration gradient. |
| cadherin binding | Binding to cadherin, a type I membrane protein involved in cell adhesion. |
| CTPase activity | Catalysis of the reaction: CTP + H2O = CDP + H+ + phosphate. May or may not be coupled to another reaction. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA helicase activity | Unwinding of a DNA helix, driven by ATP hydrolysis. |
| eukaryotic initiation factor 4E binding | Binding to eukaryotic initiation factor 4E, a polypeptide factor involved in the initiation of ribosome-mediated translation. |
| gamma-tubulin binding | Binding to the microtubule constituent protein gamma-tubulin. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
| mRNA 5'-UTR binding | Binding to an mRNA molecule at its 5' untranslated region. |
| mRNA binding | Binding to messenger RNA (mRNA), an intermediate molecule between DNA and protein. mRNA includes UTR and coding sequences, but does not contain introns. |
| nucleoside-triphosphatase activity | Catalysis of the reaction: a ribonucleoside triphosphate + H2O = a ribonucleoside diphosphate + H+ + phosphate. |
| poly(A) binding | Binding to a sequence of adenylyl residues in an RNA molecule, such as the poly(A) tail, a sequence of adenylyl residues at the 3' end of eukaryotic mRNA. |
| protein serine/threonine kinase activator activity | Binds to and increases the activity of a protein serine/threonine kinase. |
| ribosomal small subunit binding | Binding to a small ribosomal subunit. |
| RNA binding | Binding to an RNA molecule or a portion thereof. |
| RNA helicase activity | Unwinding of an RNA helix, driven by ATP hydrolysis. |
| RNA stem-loop binding | Binding to a stem-loop in an RNA molecule. An RNA stem-loop is a secondary RNA structure consisting of a double-stranded RNA (dsRNA) stem and a terminal loop. |
| RNA strand annealing activity | An activity that facilitates the formation of a complementary double-stranded RNA molecule. |
| transcription factor binding | Binding to a transcription factor, a protein required to initiate or regulate transcription. |
| translation initiation factor binding | Binding to a translation initiation factor, any polypeptide factor involved in the initiation of ribosome-mediated translation. |
49 GO annotations of biological process
| Name | Definition |
|---|---|
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cellular response to arsenic-containing substance | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of an arsenic stimulus from compounds containing arsenic, including arsenates, arsenites, and arsenides. |
| cellular response to osmotic stress | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus indicating an increase or decrease in the concentration of solutes outside the organism or cell. |
| cellular response to virus | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a virus. |
| chromosome segregation | The process in which genetic material, in the form of chromosomes, is organized into specific structures and then physically separated and apportioned to two or more sets. In eukaryotes, chromosome segregation begins with the condensation of chromosomes, includes chromosome separation, and ends when chromosomes have completed movement to the spindle poles. |
| extrinsic apoptotic signaling pathway via death domain receptors | The series of molecular signals in which a signal is conveyed from the cell surface to trigger the apoptotic death of a cell. The pathway starts with a ligand binding to a death domain receptor on the cell surface, and ends when the execution phase of apoptosis is triggered. |
| gamete generation | The generation and maintenance of gametes in a multicellular organism. A gamete is a haploid reproductive cell. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| intrinsic apoptotic signaling pathway | The series of molecular signals in which an intracellular signal is conveyed to trigger the apoptotic death of a cell. The pathway starts with reception of an intracellular signal (e.g. DNA damage, endoplasmic reticulum stress, oxidative stress etc.), and ends when the execution phase of apoptosis is triggered. The intrinsic apoptotic signaling pathway is crucially regulated by permeabilization of the mitochondrial outer membrane (MOMP). |
| lipid homeostasis | Any process involved in the maintenance of an internal steady state of lipid within an organism or cell. |
| mature ribosome assembly | The aggregation, arrangement and bonding together of the large and small ribosomal subunits into a functional ribosome. |
| negative regulation of apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of cell death by apoptotic process. |
| negative regulation of cell growth | Any process that stops, prevents, or reduces the frequency, rate, extent or direction of cell growth. |
| negative regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that stops, prevents, or reduces the frequency, rate or extent of a cysteine-type endopeptidase activity involved in the apoptotic process. |
| negative regulation of extrinsic apoptotic signaling pathway via death domain receptors | Any process that stops, prevents or reduces the frequency, rate or extent of extrinsic apoptotic signaling pathway via death domain receptors. |
| negative regulation of intrinsic apoptotic signaling pathway | Any process that stops, prevents or reduces the frequency, rate or extent of intrinsic apoptotic signaling pathway. |
| negative regulation of NIK/NF-kappaB signaling | Any process that stops, prevents or reduces the frequency, rate or extent of NIK/NF-kappaB signaling. |
| negative regulation of protein-containing complex assembly | Any process that stops, prevents, or reduces the frequency, rate or extent of protein complex assembly. |
| negative regulation of translation | Any process that stops, prevents, or reduces the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| positive regulation of apoptotic process | Any process that activates or increases the frequency, rate or extent of cell death by apoptotic process. |
| positive regulation of canonical Wnt signaling pathway | Any process that increases the rate, frequency, or extent of the Wnt signaling pathway through beta-catenin, the series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell, followed by propagation of the signal via beta-catenin, and ending with a change in transcription of target genes. |
| positive regulation of cell growth | Any process that activates or increases the frequency, rate, extent or direction of cell growth. |
| positive regulation of chemokine (C-C motif) ligand 5 production | Any process that activates or increases the frequency, rate, or extent of production of chemokine (C-C motif) ligand 5. |
| positive regulation of cysteine-type endopeptidase activity involved in apoptotic process | Any process that activates or increases the activity of a cysteine-type endopeptidase involved in the apoptotic process. |
| positive regulation of G1/S transition of mitotic cell cycle | Any signalling pathway that increases or activates a cell cycle cyclin-dependent protein kinase to modulate the switch from G1 phase to S phase of the mitotic cell cycle. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of interferon-alpha production | Any process that activates or increases the frequency, rate, or extent of interferon-alpha production. |
| positive regulation of interferon-beta production | Any process that activates or increases the frequency, rate, or extent of interferon-beta production. |
| positive regulation of NIK/NF-kappaB signaling | Any process that activates or increases the frequency, rate or extent of NIK/NF-kappaB signaling. |
| positive regulation of NLRP3 inflammasome complex assembly | Any process that activates or increases the frequency, rate or extent of NLRP3 inflammasome complex assembly. |
| positive regulation of protein acetylation | Any process that activates or increases the frequency, rate or extent of protein acetylation. |
| positive regulation of protein autophosphorylation | Any process that activates or increases the frequency, rate or extent of the phosphorylation by a protein of one or more of its own residues. |
| positive regulation of protein K63-linked ubiquitination | Any process that activates or increases the frequency, rate or extent of protein K63-linked ubiquitination. |
| positive regulation of protein serine/threonine kinase activity | Any process that increases the rate, frequency, or extent of protein serine/threonine kinase activity. |
| positive regulation of toll-like receptor 7 signaling pathway | Any process that activates or increases the frequency, rate, or extent of toll-like receptor 7 signaling pathway. |
| positive regulation of toll-like receptor 8 signaling pathway | Any process that activates or increases the frequency, rate, or extent of toll-like receptor 8 signaling pathway. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| positive regulation of translation | Any process that activates or increases the frequency, rate or extent of the chemical reactions and pathways resulting in the formation of proteins by the translation of mRNA or circRNA. |
| positive regulation of translation in response to endoplasmic reticulum stress | Any process that activates, or increases the frequency, rate or extent of translation as a result of endoplasmic reticulum stress. |
| positive regulation of translational initiation | Any process that activates or increases the frequency, rate or extent of translational initiation. |
| positive regulation of viral genome replication | Any process that activates or increases the frequency, rate or extent of viral genome replication. |
| primary miRNA processing | A process involved in the conversion of a primary microRNA transcript into a pre-microRNA molecule. |
| protein localization to cytoplasmic stress granule | A process in which a protein is transported to, or maintained in, a location within a cytoplasmic stress granule. |
| response to virus | Any process that results in a change in state or activity of a cell or an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus from a virus. |
| RNA secondary structure unwinding | The process in which a secondary structure of RNA are broken or 'melted'. |
| stress granule assembly | The aggregation, arrangement and bonding together of proteins and RNA molecules to form a stress granule. |
| translational initiation | The process preceding formation of the peptide bond between the first two amino acids of a protein. This includes the formation of a complex of the ribosome, mRNA or circRNA, and an initiation complex that contains the first aminoacyl-tRNA. |
| Wnt signaling pathway | The series of molecular signals initiated by binding of a Wnt protein to a frizzled family receptor on the surface of the target cell and ending with a change in cell state. |
25 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q5W5U4 | DDX4 | Probable ATP-dependent RNA helicase DDX4 | Bos taurus (Bovine) | SS |
| Q6GVM6 | DDX3Y | ATP-dependent RNA helicase DDX3Y | Pan troglodytes (Chimpanzee) | SS |
| P38919 | EIF4A3 | Eukaryotic initiation factor 4A-III | Homo sapiens (Human) | PR |
| Q9NQI0 | DDX4 | Probable ATP-dependent RNA helicase DDX4 | Homo sapiens (Human) | SS |
| O15523 | DDX3Y | ATP-dependent RNA helicase DDX3Y | Homo sapiens (Human) | SS |
| Q9UHL0 | DDX25 | ATP-dependent RNA helicase DDX25 | Homo sapiens (Human) | PR |
| Q9UMR2 | DDX19B | ATP-dependent RNA helicase DDX19B | Homo sapiens (Human) | PR |
| Q9NUU7 | DDX19A | ATP-dependent RNA helicase DDX19A | Homo sapiens (Human) | PR |
| P17844 | DDX5 | Probable ATP-dependent RNA helicase DDX5 | Homo sapiens (Human) | PR |
| O00148 | DDX39A | ATP-dependent RNA helicase DDX39A | Homo sapiens (Human) | PR |
| Q13838 | DDX39B | Spliceosome RNA helicase DDX39B | Homo sapiens (Human) | PR |
| Q9UJV9 | DDX41 | Probable ATP-dependent RNA helicase DDX41 | Homo sapiens (Human) | PR |
| Q61496 | Ddx4 | ATP-dependent RNA helicase DDX4 | Mus musculus (Mouse) | SS |
| Q62095 | Ddx3y | ATP-dependent RNA helicase DDX3Y | Mus musculus (Mouse) | SS |
| P16381 | D1Pas1 | Putative ATP-dependent RNA helicase Pl10 | Mus musculus (Mouse) | SS |
| Q62167 | Ddx3x | ATP-dependent RNA helicase DDX3X | Mus musculus (Mouse) | SS |
| Q6GWX0 | DDX4 | Probable ATP-dependent RNA helicase DDX4 | Sus scrofa (Pig) | SS |
| Q6Z4K6 | PL10B | DEAD-box ATP-dependent RNA helicase 52B | Oryza sativa subsp. japonica (Rice) | SS |
| Q75HJ0 | PL10A | DEAD-box ATP-dependent RNA helicase 37 | Oryza sativa subsp. japonica (Rice) | SS |
| Q2R1M8 | Os11g0599500 | DEAD-box ATP-dependent RNA helicase 52C | Oryza sativa subsp. japonica (Rice) | SS |
| O01836 | glh-3 | ATP-dependent RNA helicase glh-3 | Caenorhabditis elegans | PR |
| D0PV95 | laf-1 | ATP-dependent RNA helicase laf-1 | Caenorhabditis elegans | SS |
| Q8LA13 | RH11 | DEAD-box ATP-dependent RNA helicase 11 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q84W89 | RH37 | DEAD-box ATP-dependent RNA helicase 37 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9M2F9 | RH52 | DEAD-box ATP-dependent RNA helicase 52 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSHVAVENAL | GLDQQFAGLD | LNSSDNQSGG | STASKGRYIP | PHLRNREATK | GFYDKDSSGW |
| 70 | 80 | 90 | 100 | 110 | 120 |
| SSSKDKDAYS | SFGSRSDSRG | KSSFFSDRGS | GSRGRFDDRG | RSDYDGIGSR | GDRSGFGKFE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| RGGNSRWCDK | SDEDDWSKPL | PPSERLEQEL | FSGGNTGINF | EKYDDIPVEA | TGNNCPPHIE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SFSDVEMGEI | IMGNIELTRY | TRPTPVQKHA | IPIIKEKRDL | MACAQTGSGK | TAAFLLPILS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| QIYSDGPGEA | LRAMKENGRY | GRRKQYPISL | VLAPTRELAV | QIYEEARKFS | YRSRVRPCVV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YGGADIGQQI | RDLERGCHLL | VATPGRLVDM | MERGKIGLDF | CKYLVLDEAD | RMLDMGFEPQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| IRRIVEQDTM | PPKGVRHTMM | FSATFPKEIQ | MLARDFLDEY | IFLAVGRVGS | TSENITQKVV |
| 430 | 440 | 450 | 460 | 470 | 480 |
| WVEESDKRSF | LLDLLNATGK | DSLTLVFVET | KKGADSLEDF | LYHEGYACTS | IHGDRSQRDR |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EEALHQFRSG | KSPILVATAV | AARGLDISNV | KHVINFDLPS | DIEEYVHRIG | RTGRVGNLGL |
| 550 | 560 | 570 | 580 | 590 | 600 |
| ATSFFNERNI | NITKDLLDLL | VEAKQEVPSW | LENMAYEHHY | KGSSRGRSKS | SRFSGGFGAR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| DYRQSSGASS | SSFSSSRASS | SRSGGGGHGS | SRGFGGGGYG | GFYNSDGYGG | NYNSQGVDWW |
| GN |