O00470
Gene name |
MEIS1 |
Protein name |
Homeobox protein Meis1 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:4211 |
EC number |
|
Protein Class |
|
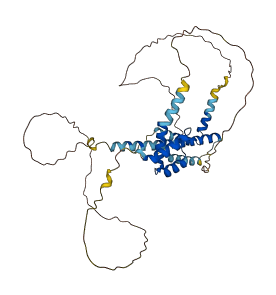
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
340-470 (C-terminal transcriptional activation domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

3 structures for O00470
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4XRS | X-ray | 350 A | A/B | 279-336 | PDB |
| 5EGO | X-ray | 254 A | A | 279-333 | PDB |
| AF-O00470-F1 | Predicted | AlphaFoldDB |
188 variants for O00470
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs371461925 CA347081742 |
4 | R>S | No |
ClinGen ESP gnomAD |
|
|
rs765836809 CA1687818 |
6 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA347081776 rs1407497093 |
6 | D>N | No |
ClinGen gnomAD |
|
|
CA347081825 rs1310247578 |
13 | G>D | No |
ClinGen gnomAD |
|
|
rs1358497771 CA347081834 |
14 | M>I | No |
ClinGen gnomAD |
|
|
CA347081832 rs1458794651 |
14 | M>T | No |
ClinGen TOPMed gnomAD |
|
|
CA347081841 rs1246939958 |
15 | D>G | No |
ClinGen gnomAD |
|
|
rs1482103507 CA347081849 |
16 | G>A | No |
ClinGen gnomAD |
|
|
CA347081853 rs1203988522 |
17 | V>L | No |
ClinGen gnomAD |
|
|
rs1573113424 CA347081861 |
18 | G>A | No |
ClinGen Ensembl |
|
|
CA1687822 rs752993508 |
19 | I>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752993508 CA347081867 |
19 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
COSM221648 CA1687823 rs758777353 |
20 | P>S | skin [Cosmic] | No |
ClinGen cosmic curated ExAC TOPMed gnomAD |
|
rs778204771 CA1687824 |
23 | M>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA347081899 rs1192346220 |
24 | Y>C | No |
ClinGen TOPMed |
|
|
CA347081911 rs1384947428 |
26 | D>A | No |
ClinGen gnomAD |
|
|
rs200427772 CA1687825 |
26 | D>E | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1384947428 CA347081913 |
26 | D>G | No |
ClinGen gnomAD |
|
|
CA347081920 rs1163790343 |
27 | P>L | No |
ClinGen gnomAD |
|
|
rs1387711149 CA347081929 |
29 | A>T | No |
ClinGen gnomAD |
|
|
CA1687827 rs781181478 |
30 | A>G | No |
ClinGen ExAC |
|
|
CA347081937 rs1169211246 |
30 | A>S | No |
ClinGen gnomAD |
|
|
CA347081935 rs1169211246 |
30 | A>T | No |
ClinGen gnomAD |
|
|
CA1687832 rs772012473 |
33 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs780426250 CA1687830 |
33 | M>L | No |
ClinGen ExAC gnomAD |
|
|
CA1687831 rs747890719 |
33 | M>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs565471424 CA1687833 |
34 | Q>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs367951046 CA1687834 |
35 | P>L | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs776061419 CA1687836 |
40 | N>K | No |
ClinGen ExAC |
|
|
rs922651749 CA347082012 |
41 | H>Q | No |
ClinGen TOPMed gnomAD |
|
|
CA50023085 rs553932802 |
43 | P>L | No |
ClinGen Ensembl |
|
|
CA1687837 rs759092516 |
45 | L>R | No |
ClinGen ExAC gnomAD |
|
|
CA347082030 rs1381550151 |
45 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1243110681 CA347082047 |
47 | S>L | No |
ClinGen gnomAD |
|
|
CA1687839 rs752369772 |
49 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs372202666 CA1687840 |
50 | Y>C | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs865922622 CA50023087 |
51 | P>Q | No |
ClinGen Ensembl |
|
|
CA347082100 rs1419561639 |
55 | H>L | No |
ClinGen gnomAD |
|
|
rs1434738144 CA347082106 |
56 | T>N | No |
ClinGen TOPMed |
|
|
rs934185879 CA50023088 |
57 | N>K | No |
ClinGen Ensembl |
|
|
rs572995046 CA50023089 |
58 | A>G | No |
ClinGen Ensembl |
|
|
rs781330399 CA1687844 |
58 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1338382686 CA347082124 |
59 | M>I | No |
ClinGen gnomAD |
|
|
CA50023090 rs985991019 |
59 | M>T | No |
ClinGen Ensembl |
|
|
CA1687846 rs545136188 |
59 | M>V | No |
ClinGen ExAC gnomAD |
|
|
CA1687847 rs780370917 |
60 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs749548319 CA1687848 |
63 | M>K | No |
ClinGen ExAC gnomAD |
|
|
rs1271023233 CA347082148 |
63 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
CA347082146 rs1271023233 |
63 | M>L | No |
ClinGen TOPMed gnomAD |
|
|
CA347082157 rs1217591995 |
64 | G>D | No |
ClinGen gnomAD |
|
|
rs954461904 CA50023091 |
65 | S>F | No |
ClinGen TOPMed gnomAD |
|
|
rs1329905600 CA347082160 |
65 | S>T | No |
ClinGen gnomAD |
|
|
rs954461904 CA347082163 |
65 | S>Y | No |
ClinGen TOPMed gnomAD |
|
|
CA50023092 rs867839014 |
67 | V>I | No |
ClinGen gnomAD |
|
|
CA50023093 rs983085178 |
68 | N>H | No |
ClinGen TOPMed gnomAD |
|
|
CA347082183 rs1266576024 |
69 | D>N | No |
ClinGen gnomAD |
|
|
CA347082193 rs1214596612 |
70 | A>S | No |
ClinGen gnomAD |
|
|
rs1259439788 CA347082194 |
70 | A>V | No |
ClinGen TOPMed |
|
|
rs866533130 CA50023094 |
72 | K>N | No |
ClinGen Ensembl |
|
|
rs1242392477 CA347082213 |
73 | R>K | No |
ClinGen gnomAD |
|
|
rs746949791 CA50023095 |
74 | D>E | No |
ClinGen ExAC gnomAD |
|
|
CA50023096 rs35067867 |
76 | D>G | No |
ClinGen Ensembl |
|
|
rs1196740598 CA347082233 |
76 | D>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1427994283 CA347082245 |
77 | A>V | No |
ClinGen gnomAD |
|
|
CA347082387 rs1573116936 |
81 | H>P | No |
ClinGen Ensembl |
|
|
rs575209192 CA50023291 |
81 | H>Q | No |
ClinGen Ensembl |
|
|
rs1462597071 CA347082390 |
82 | P>A | No |
ClinGen TOPMed gnomAD |
|
|
rs1180830551 CA347082416 |
86 | L>F | No |
ClinGen TOPMed |
|
|
CA347082495 rs1469607061 |
97 | A>T | No |
ClinGen gnomAD |
|
|
rs1458505316 CA347082499 |
98 | T>P | No |
ClinGen TOPMed |
|
|
CA347082501 rs1458505316 |
98 | T>S | No |
ClinGen TOPMed |
|
|
rs756115124 CA1687890 |
100 | T>P | No |
ClinGen ExAC gnomAD |
|
|
CA347082522 rs959662082 |
101 | P>L | No |
ClinGen TOPMed |
|
|
CA50023294 rs959662082 |
101 | P>R | No |
ClinGen TOPMed |
|
|
rs779973733 CA1687891 |
104 | P>R | No |
ClinGen ExAC gnomAD |
|
|
CA347082544 rs1321255208 |
105 | G>E | No |
ClinGen gnomAD |
|
|
rs778385393 CA1687894 |
105 | G>R | No |
ClinGen ExAC gnomAD |
|
|
rs747871060 CA1687895 |
106 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs747871060 CA347082547 |
106 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1573117062 CA347082574 |
110 | D>G | No |
ClinGen Ensembl |
|
|
rs202151942 CA1687900 |
118 | N>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs1480361314 CA347082653 |
121 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
rs561384504 CA50023295 |
122 | A>V | No |
ClinGen Ensembl |
|
|
CA50023296 rs924602028 |
123 | V>A | No |
ClinGen Ensembl |
|
|
CA347082674 rs752841255 |
125 | A>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs752841255 CA1687903 |
125 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA1687971 rs770272549 |
129 | R>L | No |
ClinGen ExAC |
|
|
rs749342831 CA1687973 |
133 | P>T | No |
ClinGen ExAC gnomAD |
|
|
CA50023368 rs749040629 |
134 | L>P | No |
ClinGen Ensembl |
|
|
CA50023370 rs978219405 |
138 | N>Y | No |
ClinGen TOPMed |
|
|
CA50023371 rs866102870 |
142 | D>N | No |
ClinGen Ensembl |
|
|
rs1183259682 CA347082832 |
145 | M>L | No |
ClinGen gnomAD |
|
|
CA50023438 rs952750461 |
146 | I>V | No |
ClinGen TOPMed gnomAD |
|
|
rs530426329 CA1688004 |
147 | Q>K | No |
ClinGen ExAC gnomAD |
|
|
CA347082910 rs983953932 |
156 | L>V | No |
ClinGen TOPMed gnomAD |
|
|
rs1458376080 CA347083013 |
168 | N>S | No |
ClinGen TOPMed |
|
|
CA347083044 rs1262513073 |
172 | R>Q | No |
ClinGen gnomAD |
|
|
rs1157772342 CA347083063 |
175 | S>C | No |
ClinGen TOPMed |
|
|
rs560708954 CA50023614 |
175 | S>N | No |
ClinGen Ensembl |
|
|
CA1688027 rs760299731 |
184 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs772211406 CA1688028 |
187 | I>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs755348268 CA1688030 |
189 | D>N | No |
ClinGen ExAC gnomAD |
|
|
rs753232642 CA1688032 |
190 | R>G | No |
ClinGen ExAC gnomAD |
|
|
rs758451287 CA1688034 |
190 | R>K | No |
ClinGen ExAC |
|
|
CA347083191 rs1400145161 |
193 | G>A | No |
ClinGen gnomAD |
|
|
rs777773287 CA1688035 |
198 | S>R | No |
ClinGen ExAC gnomAD |
|
|
CA347083249 rs1301661620 |
201 | I>R | No |
ClinGen gnomAD |
|
|
CA347083245 rs1386155008 |
201 | I>V | No |
ClinGen gnomAD |
|
|
CA50023616 rs892646699 |
202 | T>A | No |
ClinGen Ensembl |
|
|
rs369833361 CA50023617 |
203 | R>G | No |
ClinGen ESP gnomAD |
|
|
rs552329583 CA1688036 |
204 | S>T | No |
ClinGen ExAC gnomAD |
|
|
rs1285636755 CA347083277 |
206 | N>S | No |
ClinGen gnomAD |
|
|
rs554620476 CA1688037 |
208 | T>A | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1244933999 CA347083297 |
209 | D>E | No |
ClinGen TOPMed |
|
|
CA347083296 rs1244933999 |
209 | D>E | No |
ClinGen TOPMed |
|
|
rs781493613 CA1688038 |
209 | D>N | No |
ClinGen ExAC gnomAD |
|
|
CA347083300 rs1289683834 |
210 | Q>E | No |
ClinGen gnomAD |
|
|
rs760717673 CA1688039 |
210 | Q>R | No |
ClinGen ExAC gnomAD |
|
|
rs1394137805 CA347083413 |
211 | P>S | No |
ClinGen TOPMed gnomAD |
|
|
CA347083425 rs1487044408 |
213 | W>G | No |
ClinGen gnomAD |
|
|
CA1688072 rs759245209 |
217 | H>D | No |
ClinGen ExAC gnomAD |
|
|
rs1051865597 CA50032007 |
217 | H>Q | No |
ClinGen TOPMed |
|
|
CA1688074 rs376358636 |
220 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1415220775 CA347083485 |
221 | A>V | No |
ClinGen gnomAD |
|
|
CA347083493 rs1358679652 |
222 | S>F | No |
ClinGen gnomAD |
|
|
rs1399537292 CA347083498 |
223 | T>S | No |
ClinGen gnomAD |
|
|
rs867248211 CA50032010 |
224 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs867248211 CA347083501 |
224 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA1688077 COSM1214899 rs199516222 |
224 | R>H | large_intestine [Cosmic] | No |
ClinGen cosmic curated ESP ExAC TOPMed gnomAD |
|
rs1368786744 CA347083514 |
226 | G>A | No |
ClinGen gnomAD |
|
|
CA347083519 rs1280357271 |
227 | G>E | No |
ClinGen gnomAD |
|
|
rs767689616 CA347083556 |
233 | S>I | No |
ClinGen ExAC gnomAD |
|
|
CA1688079 rs767689616 |
233 | S>T | No |
ClinGen ExAC gnomAD |
|
|
CA1688081 rs546155412 |
234 | G>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs780426099 CA1688082 |
234 | G>V | No |
ClinGen ExAC gnomAD |
|
|
rs562863229 CA1688083 |
235 | G>S | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA347083572 rs1419330027 |
236 | H>L | No |
ClinGen gnomAD |
|
|
CA1688085 rs758233459 |
237 | T>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA50032017 rs531297783 |
239 | H>Y | No |
ClinGen Ensembl |
|
|
CA347083598 rs1404054563 |
240 | S>T | No |
ClinGen gnomAD |
|
|
rs1439134180 CA347083618 |
243 | N>Y | No |
ClinGen gnomAD |
|
|
CA347083646 COSM3714222 rs1135875 |
246 | E>D | upper_aerodigestive_tract [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ESP ExAC TOPMed gnomAD |
|
rs550508376 CA1688089 |
246 | E>V | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs1290610580 CA347085226 |
248 | G>D | No |
ClinGen gnomAD |
|
|
rs1158933133 CA347085240 |
250 | G>D | No |
ClinGen TOPMed |
|
|
rs61752692 CA1688120 |
254 | S>G | No |
ClinGen ExAC gnomAD |
|
|
CA347085273 rs1386075568 |
255 | V>I | No |
ClinGen gnomAD |
|
|
rs1418535282 CA347085291 |
258 | P>S | No |
ClinGen TOPMed |
|
|
rs1433718684 CA347085339 |
264 | D>E | No |
ClinGen TOPMed |
|
|
CA347085349 rs1266332568 |
266 | P>T | No |
ClinGen TOPMed |
|
|
rs751312385 CA1688125 |
272 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
VAR_063166 rs61752693 CA1688126 |
272 | R>H | found in a patient with susceptibility to restless legs syndrome [UniProt] | No |
ClinGen UniProt 1000Genomes ESP ExAC TOPMed dbSNP gnomAD |
|
rs751312385 CA1688124 |
272 | R>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs913781723 CA50041801 |
276 | R>H | No |
ClinGen Ensembl |
|
|
CA1688127 rs750403096 |
278 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA1688131 rs754588629 |
282 | V>L | No |
ClinGen ExAC gnomAD |
|
|
rs1257858865 COSM371379 CA347083348 |
289 | A>V | lung [Cosmic] | No |
ClinGen cosmic curated TOPMed |
|
rs1176438845 CA347083365 |
292 | F>V | No |
ClinGen gnomAD |
|
|
rs1218336141 CA347083812 |
317 | L>I | No |
ClinGen gnomAD |
|
|
rs1259130203 CA347083819 |
318 | Q>E | No |
ClinGen gnomAD |
|
|
CA347083818 rs1259130203 |
318 | Q>K | No |
ClinGen gnomAD |
|
|
CA347083834 rs1235206549 |
320 | N>Y | No |
ClinGen gnomAD |
|
|
rs373420611 CA50053494 |
334 | M>V | No |
ClinGen ESP TOPMed |
|
|
CA50053496 rs926938618 |
338 | S>A | No |
ClinGen TOPMed |
|
|
CA50053497 rs958348230 |
338 | S>F | No |
ClinGen TOPMed |
|
|
CA347083990 rs1345250115 |
339 | N>S | No |
ClinGen gnomAD |
|
|
rs200602768 CA1688218 |
346 | T>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA347084262 rs1278116603 |
352 | G>E | No |
ClinGen TOPMed |
|
|
CA50053708 rs547262794 |
353 | Q>H | No |
ClinGen TOPMed gnomAD |
|
|
CA1688221 rs758483192 |
355 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs758483192 CA347084284 |
355 | M>I | No |
ClinGen ExAC gnomAD |
|
|
rs1357930820 CA347084277 |
355 | M>L | No |
ClinGen TOPMed |
|
|
rs371528287 CA1688222 |
357 | G>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs2901863 CA50053711 |
358 | F>V | No |
ClinGen Ensembl |
|
|
CA1688223 rs144778967 |
359 | V>I | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs769670616 CA1688224 |
360 | M>I | No |
ClinGen ExAC gnomAD |
|
|
CA347084312 rs1276022900 |
360 | M>T | No |
ClinGen gnomAD |
|
|
rs373115262 CA50053717 |
361 | D>N | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA347084347 rs1343143228 |
365 | H>Y | No |
ClinGen gnomAD |
|
|
CA1688226 rs570343772 |
366 | M>L | No |
ClinGen ExAC gnomAD |
|
|
rs879112620 CA50053720 |
371 | P>A | No |
ClinGen Ensembl |
|
|
rs904457402 CA50053722 |
371 | P>Q | No |
ClinGen Ensembl |
|
|
CA1688243 rs755959089 |
373 | P>S | No |
ClinGen ExAC gnomAD |
|
|
CA1688245 rs373920993 |
374 | M>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA50053788 rs753394170 |
376 | G>A | No |
ClinGen Ensembl |
|
|
CA347084500 rs1455640713 |
378 | G>V | No |
ClinGen TOPMed |
|
|
rs1209337277 CA347084512 |
380 | N>H | No |
ClinGen TOPMed |
|
|
COSM1532357 rs1476459091 CA347084524 |
381 | M>I | lung [Cosmic] | No |
ClinGen cosmic curated gnomAD |
1 associated diseases with O00470
[MIM: 612853]: Restless legs syndrome 7 (RLS7)
A neurologic sleep/wake disorder characterized by uncomfortable and unpleasant sensations in the legs that appear at rest, usually at night, inducing an irresistible desire to move the legs. The disorder results in nocturnal insomnia and chronic sleep deprivation. The majority of patients also have periodic limb movements in sleep, which are characterized by involuntary, highly stereotypical, regularly occurring limb movements that occur during sleep. {ECO:0000305|PubMed:19620614}. Note=Disease susceptibility may be associated with variants affecting the gene represented in this entry.
Without disease ID
- A neurologic sleep/wake disorder characterized by uncomfortable and unpleasant sensations in the legs that appear at rest, usually at night, inducing an irresistible desire to move the legs. The disorder results in nocturnal insomnia and chronic sleep deprivation. The majority of patients also have periodic limb movements in sleep, which are characterized by involuntary, highly stereotypical, regularly occurring limb movements that occur during sleep. {ECO:0000305|PubMed:19620614}. Note=Disease susceptibility may be associated with variants affecting the gene represented in this entry.
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| chromatin | The ordered and organized complex of DNA, protein, and sometimes RNA, that forms the chromosome. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| transcription regulator complex | A protein complex that is capable of associating with DNA by direct binding, or via other DNA-binding proteins or complexes, and regulating transcription. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| DNA binding | Any molecular function by which a gene product interacts selectively and non-covalently with DNA (deoxyribonucleic acid). |
| DNA-binding transcription activator activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that activates or increases transcription of specific gene sets transcribed by RNA polymerase II. |
| DNA-binding transcription factor activity, RNA polymerase II-specific | A DNA-binding transcription factor activity that modulates the transcription of specific gene sets transcribed by RNA polymerase II. |
| RNA polymerase II cis-regulatory region sequence-specific DNA binding | Binding to a specific upstream regulatory DNA sequence (transcription factor recognition sequence or binding site) located in cis relative to the transcription start site (i.e., on the same strand of DNA) of a gene transcribed by RNA polymerase II. |
15 GO annotations of biological process
| Name | Definition |
|---|---|
| angiogenesis | Blood vessel formation when new vessels emerge from the proliferation of pre-existing blood vessels. |
| animal organ morphogenesis | Morphogenesis of an animal organ. An organ is defined as a tissue or set of tissues that work together to perform a specific function or functions. Morphogenesis is the process in which anatomical structures are generated and organized. Organs are commonly observed as visibly distinct structures, but may also exist as loosely associated clusters of cells that work together to perform a specific function or functions. |
| brain development | The process whose specific outcome is the progression of the brain over time, from its formation to the mature structure. Brain development begins with patterning events in the neural tube and ends with the mature structure that is the center of thought and emotion. The brain is responsible for the coordination and control of bodily activities and the interpretation of information from the senses (sight, hearing, smell, etc.). |
| definitive hemopoiesis | A second wave of blood cell production that, in vertebrates, generates long-term hemopoietic stem cells that continously provide erythroid, myeloid and lymphoid lineages throughout adulthood. |
| embryonic pattern specification | The process that results in the patterns of cell differentiation that will arise in an embryo. |
| eye development | The process whose specific outcome is the progression of the eye over time, from its formation to the mature structure. The eye is the organ of sight. |
| hemopoiesis | The process whose specific outcome is the progression of the myeloid and lymphoid derived organ/tissue systems of the blood and other parts of the body over time, from formation to the mature structure. The site of hemopoiesis is variable during development, but occurs primarily in bone marrow or kidney in many adult vertebrates. |
| lens morphogenesis in camera-type eye | The process in which the anatomical structures of the lens are generated and organized. The lens is a transparent structure in the eye through which light is focused onto the retina. An example of this process is found in Mus musculus. |
| locomotory behavior | The specific movement from place to place of an organism in response to external or internal stimuli. Locomotion of a whole organism in a manner dependent upon some combination of that organism's internal state and external conditions. |
| megakaryocyte development | The process whose specific outcome is the progression of a megakaryocyte cell over time, from its formation to the mature structure. Megakaryocyte development does not include the steps involved in committing a cell to a megakaryocyte fate. A megakaryocyte is a giant cell 50 to 100 micron in diameter, with a greatly lobulated nucleus, found in the bone marrow. |
| negative regulation of myeloid cell differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of myeloid cell differentiation. |
| negative regulation of neuron differentiation | Any process that stops, prevents, or reduces the frequency, rate or extent of neuron differentiation. |
| positive regulation of cell population proliferation | Any process that activates or increases the rate or extent of cell proliferation. |
| positive regulation of transcription by RNA polymerase II | Any process that activates or increases the frequency, rate or extent of transcription from an RNA polymerase II promoter. |
| regulation of transcription by RNA polymerase II | Any process that modulates the frequency, rate or extent of transcription mediated by RNA polymerase II. |
14 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q8MIB7 | TGIF2LX | Homeobox protein TGIF2LX | Pan troglodytes (Chimpanzee) | PR |
| P40424 | PBX1 | Pre-B-cell leukemia transcription factor 1 | Homo sapiens (Human) | PR |
| P40425 | PBX2 | Pre-B-cell leukemia transcription factor 2 | Homo sapiens (Human) | PR |
| Q8IUE1 | TGIF2LX | Homeobox protein TGIF2LX | Homo sapiens (Human) | PR |
| O14770 | MEIS2 | Homeobox protein Meis2 | Homo sapiens (Human) | EV |
| Q99687 | MEIS3 | Homeobox protein Meis3 | Homo sapiens (Human) | SS |
| A8K0S8 | MEIS3P2 | Putative homeobox protein Meis3-like 2 | Homo sapiens (Human) | PR |
| A6NDR6 | MEIS3P1 | Putative homeobox protein Meis3-like 1 | Homo sapiens (Human) | PR |
| P97368 | Meis3 | Homeobox protein Meis3 | Mus musculus (Mouse) | PR |
| P97367 | Meis2 | Homeobox protein Meis2 | Mus musculus (Mouse) | SS |
| Q75LX9 | Os03g0673500 | Putative homeobox protein knotted-1-like 5 | Oryza sativa subsp japonica (Rice) | PR |
| Q75LX7 | OSH10 | Homeobox protein knotted-1-like 4 | Oryza sativa subsp japonica (Rice) | PR |
| Q1PFD1 | BLH11 | BEL1-like homeodomain protein 11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SIW1 | BLH7 | BEL1-like homeodomain protein 7 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MAQRYDDLPH | YGGMDGVGIP | STMYGDPHAA | RSMQPVHHLN | HGPPLHSHQY | PHTAHTNAMA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| PSMGSSVNDA | LKRDKDAIYG | HPLFPLLALI | FEKCELATCT | PREPGVAGGD | VCSSESFNED |
| 130 | 140 | 150 | 160 | 170 | 180 |
| IAVFAKQIRA | EKPLFSSNPE | LDNLMIQAIQ | VLRFHLLELE | KVHELCDNFC | HRYISCLKGK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| MPIDLVIDDR | EGGSKSDSED | ITRSANLTDQ | PSWNRDHDDT | ASTRSGGTPG | PSSGGHTSHS |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GDNSSEQGDG | LDNSVASPST | GDDDDPDKDK | KRHKKRGIFP | KVATNIMRAW | LFQHLTHPYP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SEEQKKQLAQ | DTGLTILQVN | NWFINARRRI | VQPMIDQSNR | AVSQGTPYNP | DGQPMGGFVM |
| 370 | 380 | ||||
| DGQQHMGIRA | PGPMSGMGMN | MGMEGQWHYM |