G5EED4
Gene name |
nipi-3 |
Protein name |
Protein nipi-3 |
Names |
No induction of peptide after drechmeria infection protein 3, Tribbles homolog nipi-3 |
Species |
Caenorhabditis elegans |
KEGG Pathway |
cel:CELE_K09A9.1 |
EC number |
|
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

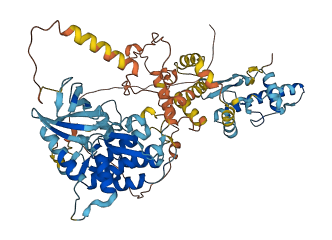
1 structures for G5EED4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-G5EED4-F1 | Predicted | AlphaFoldDB |
No variants for G5EED4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for G5EED4 | |||||
No associated diseases with G5EED4
8 regional properties for G5EED4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | SH2 domain | 280 - 376 | IPR000980 |
| domain | SH3 domain | 2 - 61 | IPR001452-1 |
| domain | SH3 domain | 106 - 165 | IPR001452-2 |
| domain | SH3 domain | 190 - 252 | IPR001452-3 |
| domain | Nck1, SH3 domain 1 | 3 - 61 | IPR035562 |
| domain | Nck1, SH3 domain 2 | 108 - 162 | IPR035564 |
| domain | Nck1, SH3 domain 3 | 193 - 249 | IPR035565 |
| domain | Nck1, SH2 domain | 280 - 376 | IPR035882 |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| mitogen-activated protein kinase kinase binding | Binding to a mitogen-activated protein kinase kinase, a protein that can phosphorylate a MAP kinase. |
| protein kinase activity | Catalysis of the phosphorylation of an amino acid residue in a protein, usually according to the reaction: a protein + ATP = a phosphoprotein + ADP. |
5 GO annotations of biological process
| Name | Definition |
|---|---|
| defense response to fungus | Reactions triggered in response to the presence of a fungus that act to protect the cell or organism. |
| positive regulation of gene expression | Any process that increases the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| positive regulation of proteasomal ubiquitin-dependent protein catabolic process | Any process that activates or increases the frequency, rate or extent of the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of MAP kinase activity | Any process that modulates the frequency, rate or extent of MAP kinase activity. |
10 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q0VCE3 | TRIB3 | Tribbles homolog 3 | Bos taurus (Bovine) | SS |
| Q5GLH2 | TRIB2 | Tribbles homolog 2 | Bos taurus (Bovine) | SS |
| Q28283 | TRIB2 | Tribbles homolog 2 | Canis lupus familiaris (Dog) (Canis familiaris) | SS |
| Q96RU8 | TRIB1 | Tribbles homolog 1 | Homo sapiens (Human) | EV |
| Q96RU7 | TRIB3 | Tribbles homolog 3 | Homo sapiens (Human) | PR |
| Q92519 | TRIB2 | Tribbles homolog 2 | Homo sapiens (Human) | SS |
| Q8K4K4 | Trib1 | Tribbles homolog 1 | Mus musculus (Mouse) | SS |
| Q8K4K2 | Trib3 | Tribbles homolog 3 | Mus musculus (Mouse) | SS |
| Q8K4K3 | Trib2 | Tribbles homolog 2 | Mus musculus (Mouse) | SS |
| Q9WTQ6 | Trib3 | Tribbles homolog 3 | Rattus norvegicus (Rat) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MARTKCKTKT | VANPRTGVRK | TAKDLSEPVR | QDAVSRRRPT | TLPFVGNTSS | RPRVFYDVLA |
| 70 | 80 | 90 | 100 | 110 | 120 |
| KKNVVFPNVK | AEYHYRVRNA | PKIDPKNFGT | TLPICYSSIG | PAKTLELRPL | GRLPPHIIKA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LNDHYVQLTR | GSMVPAADLY | NNGDHPAEQR | IPTMLNENEY | LRLIQELEEE | EKSKQFSATS |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SSAPHVNPYS | AQHLVNGEII | GIFVIYGTGL | VTRAVCSQTR | EMFTAHVLPE | WKASKVIDVI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| RRLQIPTDIS | SMTADEIRLS | ELCISKRMEI | IKSNDRYILM | NPCESATIHS | YATERLDEIT |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ENDVMSIYQK | VVEIVRFCHS | RKVILQNFKP | RSFYLKKDAH | NKWVVRPCFL | QDMSCEEDQS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| EAQFTRRSVC | VPFMAPEMLT | AESRTHHSYS | TELWGLGVLL | YILLTGKYPF | HENSMPLLFR |
| 430 | 440 | 450 | 460 | 470 | 480 |
| TIKFKQHRWP | FNFISSKSRN | IVNMLLKKAP | ATRMNLEDLW | NQVNGDFPEI | RCRSNIILKK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| QDMIVKMDLF | EMYYNTYKDR | LLPKNVLPMY | EEMKACRNDS | TILTEMAKRD | FRSIQEQMKR |
| 550 | 560 | 570 | 580 | 590 | 600 |
| RIETKPTEYQ | TLVMQVRLQQ | INQLFFEKEV | SQAQKQHRAP | RLVQLKCSDI | SKELLLPGDI |
| 610 | 620 | 630 | 640 | 650 | |
| YPISEHYHPS | QQPVDKVVYK | LLSDANSLAF | PTVMKGTVPK | SYPPPVFKGL | DISPS |