F1N9Y5
Gene name |
SYK (RCJMB04_19o18) |
Protein name |
Tyrosine-protein kinase SYK |
Names |
|
Species |
Gallus gallus (Chicken) |
KEGG Pathway |
gga:427272 |
EC number |
|
Protein Class |
|
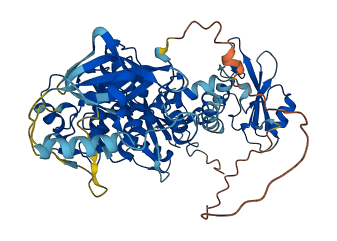
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
349-613 (Protein kinase domain) |
Relief mechanism |
Partner binding, PTM |
Assay |
|
Target domain |
349-613 (Protein kinase domain) |
Relief mechanism |
Partner binding, PTM |
Assay |
|
Accessory elements
489-515 (Activation loop from InterPro)
Target domain |
349-613 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
References
- Adachi T et al. (2007) "Interdomain A is crucial for ITAM-dependent and -independent regulation of Syk", Biochemical and biophysical research communications, 364, 111-7
- Kulathu Y et al. (2009) "Autoinhibition and adapter function of Syk", Immunological reviews, 232, 286-99
- Bond PJ et al. (2011) "Molecular mechanism of selective recruitment of Syk kinases by the membrane antigen-receptor complex", The Journal of biological chemistry, 286, 25872-81
- Brdicka T et al. (2005) "Intramolecular regulatory switch in ZAP-70: analogy with receptor tyrosine kinases", Molecular and cellular biology, 25, 4924-33
- Cai X et al. (2008) "Spatial and temporal regulation of focal adhesion kinase activity in living cells", Molecular and cellular biology, 28, 201-14
- Herzog FA et al. (2017) "Structural Insights How PIP2 Imposes Preferred Binding Orientations of FAK at Lipid Membranes", The journal of physical chemistry. B, 121, 3523-3535
- Williams JC et al. (1997) "The 2.35 A crystal structure of the inactivated form of chicken Src: a dynamic molecule with multiple regulatory interactions", Journal of molecular biology, 274, 757-75
- Boggon TJ et al. (2004) "Structure and regulation of Src family kinases", Oncogene, 23, 7918-27
- Meng Y et al. (2014) "Locking the active conformation of c-Src kinase through the phosphorylation of the activation loop", Journal of molecular biology, 426, 423-35
- Register AC et al. (2014) "SH2-catalytic domain linker heterogeneity influences allosteric coupling across the SFK family", Biochemistry, 53, 6910-23
Autoinhibited structure

Activated structure

1 structures for F1N9Y5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-F1N9Y5-F1 | Predicted | AlphaFoldDB |
No variants for F1N9Y5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for F1N9Y5 | |||||
No associated diseases with F1N9Y5
8 regional properties for F1N9Y5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 349 - 613 | IPR000719 |
| domain | SH2 domain | 12 - 106 | IPR000980-1 |
| domain | SH2 domain | 165 - 258 | IPR000980-2 |
| domain | Serine-threonine/tyrosine-protein kinase, catalytic domain | 354 - 601 | IPR001245 |
| active_site | Tyrosine-protein kinase, active site | 468 - 480 | IPR008266 |
| binding_site | Protein kinase, ATP binding site | 355 - 380 | IPR017441 |
| domain | Tyrosine-protein kinase, catalytic domain | 349 - 604 | IPR020635 |
| domain | SYK/ZAP-70, N-terminal SH2 domain | 12 - 115 | IPR035838 |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| early phagosome | A membrane-bounded intracellular vesicle as initially formed upon the ingestion of particulate material by phagocytosis. |
| extrinsic component of cytoplasmic side of plasma membrane | The component of a plasma membrane consisting of gene products and protein complexes that are loosely bound to its cytoplasmic surface, but not integrated into the hydrophobic region. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| non-membrane spanning protein tyrosine kinase activity | Catalysis of the reaction: ATP + protein L-tyrosine = ADP + protein L-tyrosine phosphate by a non-membrane spanning protein. |
| protein serine/threonine/tyrosine kinase activity | Catalysis of the reactions: ATP + a protein serine = ADP + protein serine phosphate; ATP + a protein threonine = ADP + protein threonine phosphate; and ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| protein tyrosine kinase activity | Catalysis of the reaction: ATP + a protein tyrosine = ADP + protein tyrosine phosphate. |
| signaling receptor binding | Binding to one or more specific sites on a receptor molecule, a macromolecule that undergoes combination with a hormone, neurotransmitter, drug or intracellular messenger to initiate a change in cell function. |
31 GO annotations of biological process
| Name | Definition |
|---|---|
| adaptive immune response | An immune response mediated by cells expressing specific receptors for antigen produced through a somatic diversification process, and allowing for an enhanced secondary response to subsequent exposures to the same antigen (immunological memory). |
| B cell receptor signaling pathway | The series of molecular signals initiated by the cross-linking of an antigen receptor on a B cell. |
| blood vessel morphogenesis | The process in which the anatomical structures of blood vessels are generated and organized. The blood vessel is the vasculature carrying blood. |
| cell differentiation | The process in which relatively unspecialized cells, e.g. embryonic or regenerative cells, acquire specialized structural and/or functional features that characterize the cells, tissues, or organs of the mature organism or some other relatively stable phase of the organism's life history. Differentiation includes the processes involved in commitment of a cell to a specific fate and its subsequent development to the mature state. |
| cellular response to hydrogen peroxide | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a hydrogen peroxide (H2O2) stimulus. |
| cellular response to molecule of fungal origin | Any process that results in a change in state or activity of a cell (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of fungal origin such as chito-octamer oligosaccharide. |
| defense response to bacterium | Reactions triggered in response to the presence of a bacterium that act to protect the cell or organism. |
| innate immune response | Innate immune responses are defense responses mediated by germline encoded components that directly recognize components of potential pathogens. |
| integrin-mediated signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to an integrin on the surface of a target cell, and ending with the regulation of a downstream cellular process, e.g. transcription. |
| intracellular signal transduction | The process in which a signal is passed on to downstream components within the cell, which become activated themselves to further propagate the signal and finally trigger a change in the function or state of the cell. |
| leukocyte activation involved in immune response | A change in morphology and behavior of a leukocyte resulting from exposure to a specific antigen, mitogen, cytokine, cellular ligand, or soluble factor, leading to the initiation or perpetuation of an immune response. |
| leukocyte cell-cell adhesion | The attachment of a leukocyte to another cell via adhesion molecules. |
| lymph vessel development | The process whose specific outcome is the progression of a lymph vessel over time, from its formation to the mature structure. |
| macrophage activation involved in immune response | A change in morphology and behavior of a macrophage resulting from exposure to a cytokine, chemokine, cellular ligand, or soluble factor, leading to the initiation or perpetuation of an immune response. |
| neutrophil activation involved in immune response | The change in morphology and behavior of a neutrophil resulting from exposure to a cytokine, chemokine, cellular ligand, or soluble factor, leading to the initiation or perpetuation of an immune response. |
| neutrophil chemotaxis | The directed movement of a neutrophil cell, the most numerous polymorphonuclear leukocyte found in the blood, in response to an external stimulus, usually an infection or wounding. |
| peptidyl-tyrosine phosphorylation | The phosphorylation of peptidyl-tyrosine to form peptidyl-O4'-phospho-L-tyrosine. |
| positive regulation of alpha-beta T cell proliferation | Any process that activates or increases the frequency, rate or extent of alpha-beta T cell proliferation. |
| positive regulation of B cell differentiation | Any process that activates or increases the frequency, rate or extent of B cell differentiation. |
| positive regulation of bone resorption | Any process that activates or increases the frequency, rate or extent of bone resorption. |
| positive regulation of cell adhesion mediated by integrin | Any process that activates or increases the frequency, rate, or extent of cell adhesion mediated by integrin. |
| positive regulation of mast cell degranulation | Any process that activates or increases the frequency, rate or extent of mast cell degranulation. |
| regulation of arachidonic acid secretion | Any process that modulates the rate, frequency, or extent of arachidonic acid secretion, the controlled release of arachidonic acid from a cell or a tissue. |
| regulation of ERK1 and ERK2 cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the ERK1 and ERK2 cascade. |
| regulation of neutrophil degranulation | Any process that modulates the frequency, rate, or extent of neutrophil degranulation. |
| regulation of phagocytosis | Any process that modulates the frequency, rate or extent of phagocytosis, the process in which phagocytes engulf external particulate material. |
| regulation of platelet activation | Any process that modulates the rate or frequency of platelet activation. Platelet activation is a series of progressive, overlapping events triggered by exposure of the platelets to subendothelial tissue. |
| regulation of platelet aggregation | Any process that modulates the rate, frequency or extent of platelet aggregation. Platelet aggregation is the adhesion of one platelet to one or more other platelets via adhesion molecules. |
| regulation of superoxide anion generation | Any process that modulates the frequency, rate or extent of enzymatic generation of superoxide by a cell. |
| serotonin secretion by platelet | The regulated release of serotonin by a platelet or group of platelets. |
| transmembrane receptor protein tyrosine kinase signaling pathway | The series of molecular signals initiated by an extracellular ligand binding to a receptor on the surface of the target cell where the receptor possesses tyrosine kinase activity, and ending with the regulation of a downstream cellular process, e.g. transcription. |
19 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P00523 | SRC | Proto-oncogene tyrosine-protein kinase Src | Gallus gallus (Chicken) | EV |
| Q02977 | YRK | Proto-oncogene tyrosine-protein kinase Yrk | Gallus gallus (Chicken) | SS |
| P09324 | YES1 | Tyrosine-protein kinase Yes | Gallus gallus (Chicken) | SS |
| P41239 | CSK | Tyrosine-protein kinase CSK | Gallus gallus (Chicken) | SS |
| Q00944 | PTK2 | Focal adhesion kinase 1 | Gallus gallus (Chicken) | EV |
| P42683 | LCK | Proto-oncogene tyrosine-protein kinase LCK | Gallus gallus (Chicken) | SS |
| Q8JH64 | BTK | Tyrosine-protein kinase BTK | Gallus gallus (Chicken) | SS |
| Q05876 | FYN | Tyrosine-protein kinase Fyn | Gallus gallus (Chicken) | SS |
| P43403 | ZAP70 | Tyrosine-protein kinase ZAP-70 | Homo sapiens (Human) | EV |
| P43405 | SYK | Tyrosine-protein kinase SYK | Homo sapiens (Human) | EV |
| P43404 | Zap70 | Tyrosine-protein kinase ZAP-70 | Mus musculus (Mouse) | SS |
| P48025 | Syk | Tyrosine-protein kinase SYK | Mus musculus (Mouse) | SS |
| Q00655 | SYK | Tyrosine-protein kinase SYK | Sus scrofa (Pig) | SS |
| Q64725 | Syk | Tyrosine-protein kinase SYK | Rattus norvegicus (Rat) | SS |
| Q9SYA0 | At1g61500 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g61500 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9FG33 | LECRKS5 | Probable L-type lectin-domain containing receptor kinase S.5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O22558 | STY8 | Serine/threonine-protein kinase STY8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWL6 | STY17 | Serine/threonine-protein kinase STY17 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| F4JTP5 | STY46 | Serine/threonine-protein kinase STY46 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MASNMANPAN | HLPYFFGNIT | REEAEEYLMQ | GGMSDGLYLL | RQSRNYLGGF | ALSLAYGRKV |
| 70 | 80 | 90 | 100 | 110 | 120 |
| HHYTIERELS | GTYAIAGGKS | HASPAELINY | HSEEADGLIC | LLRKSFNRPP | GVEPKTGPFE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| DLKENLIREY | VKQTWNLQGH | ALEQAIISQK | PQLEKLIATT | AHEKMPWFHG | RISREESEHR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ILIGSRNNGK | FLIRERDSNG | SYALCLLNDG | KVLHYRIDRD | KTGKLSIPDG | KRFDTLWQLV |
| 250 | 260 | 270 | 280 | 290 | 300 |
| EHYSYKPDGL | LRVLSIPCPR | HGSESDNVVF | DTRPLPGTPS | KLQTPIGAPS | DDQTPFNPYV |
| 310 | 320 | 330 | 340 | 350 | 360 |
| LQRARGLIGA | EKGDQREALP | MDTEVYESPY | ADPDEIKPKN | VTLDRKLLTL | EEGELGSGNF |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GTVKKGFYKM | KKGAKPVAVK | ILKNESNDPA | IKDELLREAN | VMQQLDNPYI | VRMIGICEAE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AWMLVMEMAE | LGPLNKFLQK | NRHVTEKNIT | ELVHQVSMGM | KYLEENNFVH | RDLAARNVLL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| VTQHYAKISD | FGLSKALSAD | ENYYKAQSHG | KWPVKWYAPE | CMNFYKFSSK | SDVWSFGVLM |
| 550 | 560 | 570 | 580 | 590 | 600 |
| WEAFSYGQKP | YKGMKGGEVA | QMIERGERME | CPEACPVEVY | DLMKLCWTYN | VDDRPGFVAV |
| 610 | |||||
| ELRLRNYYYD | ISH |