D0ZVG2
Gene name |
sspH1 (STM14_1483) |
Protein name |
E3 ubiquitin-protein ligase SspH1 |
Names |
RING-type E3 ubiquitin transferase SspH1, Salmonella secreted protein H1, Secreted effector protein SspH1 |
Species |
Salmonella typhimurium (strain 14028s / SGSC 2262) |
KEGG Pathway |
seo:STM14_1483 |
EC number |
2.3.2.27: Aminoacyltransferases |
Protein Class |
FAMILY NOT NAMED (PTHR47114) |
Descriptions
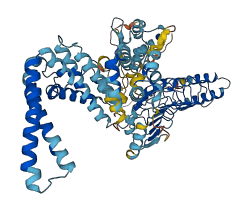
SspH1 is a Salmonella enterica serovar Typhimurium TTSS effector that localizes to the mammalian nucleus and catalyzes the ubiquitination and proteasome-dependent degradation of PKN1 in cells, which attenuates androgen receptor responsiveness. In the absence of substrate, the leucine-rich repeat (LRR) domain inhibits the catalytic domain (NEL domain). Binding of HR1b domain of PKN1 to the LRR domain relieves the autoinhibition.
Autoinhibitory domains (AIDs)
Target domain |
404-700 (E3 ubiquitin-protein ligase catalytic domain.) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Structural analysis |
Accessory elements
No accessory elements
References
- Keszei AF et al. (2014) "Structure of an SspH1-PKN1 complex reveals the basis for host substrate recognition and mechanism of activation for a bacterial E3 ubiquitin ligase", Molecular and cellular biology, 34, 362-73
- Haraga A et al. (2006) "A Salmonella type III secretion effector interacts with the mammalian serine/threonine protein kinase PKN1", Cellular microbiology, 8, 837-46
Autoinhibited structure
Activated structure
3 structures for D0ZVG2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| 4NKG | X-ray | 290 A | A/C | 161-405 | PDB |
| 4NKH | X-ray | 275 A | A/B/C/D/E/F | 161-398 | PDB |
| AF-D0ZVG2-F1 | Predicted | AlphaFoldDB |
No variants for D0ZVG2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for D0ZVG2 | |||||
No associated diseases with D0ZVG2
3 regional properties for D0ZVG2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| repeat | Leucine-rich repeat | 298 - 319 | IPR001611-1 |
| repeat | Leucine-rich repeat | 338 - 360 | IPR001611-2 |
| domain | Novel E3 ligase domain | 414 - 626 | IPR029487 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.27 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | PTHR47114 | FAMILY NOT NAMED |
| PANTHER Subfamily | PTHR47114:SF2 | OLIGODENDROCYTE-MYELIN GLYCOPROTEIN |
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular region | The space external to the outermost structure of a cell. For cells without external protective or external encapsulating structures this refers to space outside of the plasma membrane. This term covers the host cell environment outside an intracellular parasite. |
| host cell cytoplasm | The cytoplasm of a host cell. |
| host cell nucleus | A membrane-bounded organelle as it is found in the host cell in which chromosomes are housed and replicated. The host is defined as the larger of the organisms involved in a symbiotic interaction. |
1 GO annotations of molecular function
| Name | Definition |
|---|---|
| ubiquitin-protein transferase activity | Catalysis of the transfer of ubiquitin from one protein to another via the reaction X-Ub + Y --> Y-Ub + X, where both X-Ub and Y-Ub are covalent linkages. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| protein secretion by the type III secretion system | The process in which proteins are transferred into the extracellular milieu or directly into host cells by the bacterial type III secretion system; secretion occurs in a continuous process without the distinct presence of periplasmic intermediates and does not involve proteolytic processing of secreted proteins. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MFNIRNTQPS | VSMQAIAGAA | APEASPEEIV | WEKIQVFFPQ | ENYEEAQQCL | AELCHPARGM |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LPDHISSQFA | RLKALTFPAW | EENIQCNRDG | INQFCILDAG | SKEILSITLD | DAGNYTVNCQ |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GYSEAHDFIM | DTEPGEECTE | FAEGASGTSL | RPATTVSQKA | AEYDAVWSKW | ERDAPAGESP |
| 190 | 200 | 210 | 220 | 230 | 240 |
| GRAAVVQEMR | DCLNNGNPVL | NVGASGLTTL | PDRLPPHITT | LVIPDNNLTS | LPELPEGLRE |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LEVSGNLQLT | SLPSLPQGLQ | KLWAYNNWLA | SLPTLPPGLG | DLAVSNNQLT | SLPEMPPALR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ELRVSGNNLT | SLPALPSGLQ | KLWAYNNRLT | SLPEMSPGLQ | ELDVSHNQLT | RLPQSLTGLS |
| 370 | 380 | 390 | 400 | 410 | 420 |
| SAARVYLDGN | PLSVRTLQAL | RDIIGHSGIR | IHFDMAGPSV | PREARALHLA | VADWLTSARE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| GEAAQADRWQ | AFGLEDNAAA | FSLVLDRLRE | TENFKKDAGF | KAQISSWLTQ | LAEDAALRAK |
| 490 | 500 | 510 | 520 | 530 | 540 |
| TFAMATEATS | TCEDRVTHAL | HQMNNVQLVH | NAEKGEYDNN | LQGLVSTGRE | MFRLATLEQI |
| 550 | 560 | 570 | 580 | 590 | 600 |
| AREKAGTLAL | VDDVEVYLAF | QNKLKESLEL | TSVTSEMRFF | DVSGVTVSDL | QAAELQVKTA |
| 610 | 620 | 630 | 640 | 650 | 660 |
| ENSGFSKWIL | QWGPLHSVLE | RKVPERFNAL | REKQISDYED | TYRKLYDEVL | KSSGLVDDTD |
| 670 | 680 | 690 | |||
| AERTIGVSAM | DSAKKEFLDG | LRALVDEVLG | SYLTARWRLN |