C0LGT6
Gene name |
EFR |
Protein name |
LRR receptor-like serine/threonine-protein kinase EFR |
Names |
Elongation factor Tu receptor, EF-Tu receptor |
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT5G20480 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
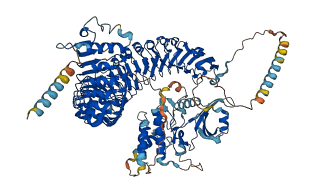
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for C0LGT6
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-C0LGT6-F1 | Predicted | AlphaFoldDB |
118 variants for C0LGT6
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| tmp_5_6922534_C_T | 13 | T>M | No | 1000Genomes | |
| tmp_5_6922551_T_A | 19 | C>S | No | 1000Genomes | |
| ENSVATH10883071 | 21 | F>S | No | 1000Genomes | |
| ENSVATH07001591 | 22 | A>S | No | 1000Genomes | |
| ENSVATH03085744 | 36 | L>V | No | 1000Genomes | |
| ENSVATH03085745 | 44 | E>G | No | 1000Genomes | |
| tmp_5_6922653_T_C | 53 | S>P | No | 1000Genomes | |
| ENSVATH00634705 | 88 | V>M | No | 1000Genomes | |
| tmp_5_6922823_T_G | 109 | F>L | No | 1000Genomes | |
| ENSVATH07001594 | 111 | S>N | No | 1000Genomes | |
| ENSVATH03085749 | 116 | K>E | No | 1000Genomes | |
| ENSVATH03085750 | 119 | R>M | No | 1000Genomes | |
| ENSVATH00634706 | 134 | E>V | No | 1000Genomes | |
| ENSVATH09576248 | 139 | S>P | No | 1000Genomes | |
| ENSVATH00634708 | 148 | S>L | No | 1000Genomes | |
| tmp_5_6922951_T_A | 152 | L>* | No | 1000Genomes | |
| tmp_5_6922971_C_G | 159 | H>D | No | 1000Genomes | |
| tmp_5_6923006_G_C | 170 | K>N | No | 1000Genomes | |
| tmp_5_6923047_A_T | 184 | N>I | No | 1000Genomes | |
| ENSVATH07001597 | 184 | N>K | No | 1000Genomes | |
| tmp_5_6923150_A_T | 218 | Q>H | No | 1000Genomes | |
| tmp_5_6923178_A_T | 228 | S>C | No | 1000Genomes | |
| ENSVATH07001601 | 235 | P>L | No | 1000Genomes | |
| tmp_5_6923229_T_A | 245 | S>T | No | 1000Genomes | |
| tmp_5_6923255_T_G | 253 | F>L | No | 1000Genomes | |
| tmp_5_6923271_G_C | 259 | A>P | No | 1000Genomes | |
| tmp_5_6923287_T_A | 264 | L>H | No | 1000Genomes | |
| ENSVATH09576258 | 270 | R>K | No | 1000Genomes | |
| ENSVATH07001602 | 270 | R>S | No | 1000Genomes | |
| tmp_5_6923332_C_G | 279 | T>S | No | 1000Genomes | |
| ENSVATH07001603 | 284 | K>I | No | 1000Genomes | |
| tmp_5_6923350_C_T | 285 | T>I | No | 1000Genomes | |
| ENSVATH07001604 | 285 | T>S | No | 1000Genomes | |
| ENSVATH10883072 | 296 | D>Y | No | 1000Genomes | |
| ENSVATH07001605 | 297 | I>T | No | 1000Genomes | |
| ENSVATH00634712 | 303 | S>T | No | 1000Genomes | |
| ENSVATH14106721 | 321 | I>S | No | 1000Genomes | |
| ENSVATH00634713 | 339 | A>P | No | 1000Genomes | |
| ENSVATH07001609 | 340 | V>L | No | 1000Genomes | |
| ENSVATH07001611 | 344 | T>N | No | 1000Genomes | |
| ENSVATH00634714 | 362 | A>D | No | 1000Genomes | |
| ENSVATH03085758 | 384 | T>N | No | 1000Genomes | |
| ENSVATH03085758 | 384 | T>S | No | 1000Genomes | |
| ENSVATH03085760 | 387 | H>R | No | 1000Genomes | |
| ENSVATH09576274 | 399 | S>N | No | 1000Genomes | |
| tmp_5_6923713_G_C | 406 | S>T | No | 1000Genomes | |
| ENSVATH14106722 | 439 | N>Y | No | 1000Genomes | |
| tmp_5_6923833_T_G | 446 | L>R | No | 1000Genomes | |
| ENSVATH00634718 | 472 | M>I | No | 1000Genomes | |
| ENSVATH08418700 | 474 | T>A | No | 1000Genomes | |
| tmp_5_6923935_C_G | 480 | T>S | No | 1000Genomes | |
| ENSVATH00634719 | 483 | Q>R | No | 1000Genomes | |
| ENSVATH00634720 | 486 | L>M | No | 1000Genomes | |
| tmp_5_6923961_C_A | 489 | P>T | No | 1000Genomes | |
| ENSVATH07001614 | 528 | K>N | No | 1000Genomes | |
| ENSVATH03085767 | 529 | M>I | No | 1000Genomes | |
| tmp_5_6924108_T_C | 538 | S>P | No | 1000Genomes | |
| ENSVATH03085768 | 543 | F>Y | No | 1000Genomes | |
| ENSVATH07001615 | 545 | Q>E | No | 1000Genomes | |
| tmp_5_6924139_C_T | 548 | S>L | No | 1000Genomes | |
| ENSVATH00634722 | 552 | A>P | No | 1000Genomes | |
| tmp_5_6924168_C_G | 558 | R>G | No | 1000Genomes | |
| tmp_5_6924169_G_A | 558 | R>Q | No | 1000Genomes | |
| tmp_5_6924183_A_G | 563 | K>E | No | 1000Genomes | |
| tmp_5_6924190_T_G | 565 | V>G | No | 1000Genomes | |
| tmp_5_6924210_C_T | 572 | L>F | No | 1000Genomes | |
| tmp_5_6924220_G_T | 575 | R>L | No | 1000Genomes | |
| tmp_5_6924225_C_T | 577 | P>S | No | 1000Genomes | |
| ENSVATH00634723 | 585 | S>L | No | 1000Genomes | |
| ENSVATH03085769 | 591 | L>I | No | 1000Genomes | |
| ENSVATH00634724 | 599 | R>M | No | 1000Genomes | |
| tmp_5_6924322_C_G | 609 | A>G | No | 1000Genomes | |
| ENSVATH07001617 | 624 | V>I | No | 1000Genomes | |
| ENSVATH07001618 | 625 | R>L | No | 1000Genomes | |
| tmp_5_6924440_G_T | 648 | K>N | No | 1000Genomes | |
| ENSVATH07001621 | 651 | V>A | No | 1000Genomes | |
| tmp_5_6924457_T_C | 654 | I>T | No | 1000Genomes | |
| tmp_5_6924472_C_G | 659 | A>G | No | 1000Genomes | |
| ENSVATH07001624 | 677 | K>R | No | 1000Genomes | |
| ENSVATH14106724 | 690 | S>F | No | 1000Genomes | |
| ENSVATH07001627 | 692 | T>I | No | 1000Genomes | |
| ENSVATH00634726 | 698 | E>K | No | 1000Genomes | |
| ENSVATH10883074 | 706 | H>L | No | 1000Genomes | |
| tmp_5_6924617_T_G | 707 | S>R | No | 1000Genomes | |
| ENSVATH07001629 | 711 | R>G | No | 1000Genomes | |
| tmp_5_6924628_G_A | 711 | R>H | No | 1000Genomes | |
| tmp_5_6924699_A_G | 735 | N>D | No | 1000Genomes | |
| tmp_5_6924855_A_G | 787 | R>G | No | 1000Genomes | |
| ENSVATH14106785 | 796 | K>N | No | 1000Genomes | |
| tmp_5_6924886_G_A | 797 | G>E | No | 1000Genomes | |
| tmp_5_6924899_G_T | 801 | M>I | No | 1000Genomes | |
| ENSVATH07001637 | 805 | L>P | No | 1000Genomes | |
| ENSVATH07001638 | 808 | L>Q | No | 1000Genomes | |
| tmp_5_6924928_T_C | 811 | V>A | No | 1000Genomes | |
| tmp_5_6924927_G_A | 811 | V>I | No | 1000Genomes | |
| ENSVATH07001639 | 813 | D>E | No | 1000Genomes | |
| tmp_5_6924940_C_T | 815 | S>L | No | 1000Genomes | |
| ENSVATH03085770 | 820 | P>L | No | 1000Genomes | |
| tmp_5_6924957_G_C | 821 | A>P | No | 1000Genomes | |
| tmp_5_6925033_C_T | 846 | A>V | No | 1000Genomes | |
| ENSVATH07001642 | 853 | S>C | No | 1000Genomes | |
| ENSVATH07001643 | 855 | I>V | No | 1000Genomes | |
| ENSVATH07001644 | 859 | D>N | No | 1000Genomes | |
| ENSVATH07001645 | 879 | R>G | No | 1000Genomes | |
| tmp_5_6925141_T_A | 882 | F>Y | No | 1000Genomes | |
| ENSVATH14106786 | 928 | K>N | No | 1000Genomes | |
| tmp_5_6925417_C_T | 945 | T>I | No | 1000Genomes | |
| tmp_5_6925441_C_T | 953 | T>M | No | 1000Genomes | |
| tmp_5_6925444_G_A | 954 | S>N | No | 1000Genomes | |
| ENSVATH07001663 | 964 | G>W | No | 1000Genomes | |
| tmp_5_6925490_G_T | 969 | L>F | No | 1000Genomes | |
| ENSVATH07001664 | 973 | I>V | No | 1000Genomes | |
| tmp_5_6925522_C_T | 980 | P>L | No | 1000Genomes | |
| ENSVATH03085773 | 993 | L>I | No | 1000Genomes | |
| tmp_5_6925590_T_C | 1003 | S>P | No | 1000Genomes | |
| ENSVATH00634728 | 1014 | A>V | No | 1000Genomes | |
| ENSVATH07001667 | 1026 | T>A | No | 1000Genomes | |
| tmp_5_6925675_T_C | 1031 | M>T | No | 1000Genomes |
No associated diseases with C0LGT6
16 regional properties for C0LGT6
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 712 - 1001 | IPR000719 |
| repeat | Leucine-rich repeat | 122 - 144 | IPR001611-1 |
| repeat | Leucine-rich repeat | 146 - 204 | IPR001611-2 |
| repeat | Leucine-rich repeat | 266 - 326 | IPR001611-3 |
| repeat | Leucine-rich repeat | 345 - 367 | IPR001611-4 |
| repeat | Leucine-rich repeat, typical subtype | 96 - 120 | IPR003591-1 |
| repeat | Leucine-rich repeat, typical subtype | 168 - 191 | IPR003591-2 |
| repeat | Leucine-rich repeat, typical subtype | 192 - 216 | IPR003591-3 |
| repeat | Leucine-rich repeat, typical subtype | 265 - 288 | IPR003591-4 |
| repeat | Leucine-rich repeat, typical subtype | 392 - 415 | IPR003591-5 |
| repeat | Leucine-rich repeat, typical subtype | 416 - 439 | IPR003591-6 |
| repeat | Leucine-rich repeat, typical subtype | 440 - 464 | IPR003591-7 |
| repeat | Leucine-rich repeat, typical subtype | 583 - 608 | IPR003591-8 |
| active_site | Serine/threonine-protein kinase, active site | 845 - 857 | IPR008271 |
| domain | Leucine-rich repeat-containing N-terminal, plant-type | 29 - 68 | IPR013210 |
| binding_site | Protein kinase, ATP binding site | 718 - 741 | IPR017441 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
3 GO annotations of cellular component
| Name | Definition |
|---|---|
| endomembrane system | A collection of membranous structures involved in transport within the cell. The main components of the endomembrane system are endoplasmic reticulum, Golgi bodies, vesicles, cell membrane and nuclear envelope. Members of the endomembrane system pass materials through each other or though the use of vesicles. |
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
4 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
| transmembrane receptor protein kinase activity | Combining with a signal and transmitting the signal from one side of the membrane to the other to initiate a change in cell activity by catalysis of the reaction: a protein + ATP = a phosphoprotein + ADP. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| detection of bacterium | The series of events in which a stimulus from a bacterium is received and converted into a molecular signal. |
| hormone-mediated signaling pathway | The series of molecular signals mediated by the detection of a hormone. |
| immune response-regulating signaling pathway | The cascade of processes by which a signal interacts with a receptor, causing a change in the level or activity of a second messenger or other downstream target, and ultimately leading to the activation, perpetuation, or inhibition of an immune response. |
| plant-type hypersensitive response | The rapid, localized death of plant cells in response to invasion by a pathogen. |
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
| regulation of anion channel activity | Any process that modulates the frequency, rate or extent of anion channel activity. |
| response to molecule of bacterial origin | Any process that results in a change in state or activity of an organism (in terms of movement, secretion, enzyme production, gene expression, etc.) as a result of a stimulus by molecules of bacterial origin such as peptides derived from bacterial flagellin. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| C0LGE4 | At1g12460 | Probable LRR receptor-like serine/threonine-protein kinase At1g12460 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGJ1 | At1g74360 | Probable LRR receptor-like serine/threonine-protein kinase At1g74360 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6XAT2 | ERL2 | LRR receptor-like serine/threonine-protein kinase ERL2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SHI4 | RLP3 | Receptor-like protein 3 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MKLSFSLVFN | ALTLLLQVCI | FAQARFSNET | DMQALLEFKS | QVSENNKREV | LASWNHSSPF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| CNWIGVTCGR | RRERVISLNL | GGFKLTGVIS | PSIGNLSFLR | LLNLADNSFG | STIPQKVGRL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| FRLQYLNMSY | NLLEGRIPSS | LSNCSRLSTV | DLSSNHLGHG | VPSELGSLSK | LAILDLSKNN |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LTGNFPASLG | NLTSLQKLDF | AYNQMRGEIP | DEVARLTQMV | FFQIALNSFS | GGFPPALYNI |
| 250 | 260 | 270 | 280 | 290 | 300 |
| SSLESLSLAD | NSFSGNLRAD | FGYLLPNLRR | LLLGTNQFTG | AIPKTLANIS | SLERFDISSN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| YLSGSIPLSF | GKLRNLWWLG | IRNNSLGNNS | SSGLEFIGAV | ANCTQLEYLD | VGYNRLGGEL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| PASIANLSTT | LTSLFLGQNL | ISGTIPHDIG | NLVSLQELSL | ETNMLSGELP | VSFGKLLNLQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| VVDLYSNAIS | GEIPSYFGNM | TRLQKLHLNS | NSFHGRIPQS | LGRCRYLLDL | WMDTNRLNGT |
| 490 | 500 | 510 | 520 | 530 | 540 |
| IPQEILQIPS | LAYIDLSNNF | LTGHFPEEVG | KLELLVGLGA | SYNKLSGKMP | QAIGGCLSME |
| 550 | 560 | 570 | 580 | 590 | 600 |
| FLFMQGNSFD | GAIPDISRLV | SLKNVDFSNN | NLSGRIPRYL | ASLPSLRNLN | LSMNKFEGRV |
| 610 | 620 | 630 | 640 | 650 | 660 |
| PTTGVFRNAT | AVSVFGNTNI | CGGVREMQLK | PCIVQASPRK | RKPLSVRKKV | VSGICIGIAS |
| 670 | 680 | 690 | 700 | 710 | 720 |
| LLLIIIVASL | CWFMKRKKKN | NASDGNPSDS | TTLGMFHEKV | SYEELHSATS | RFSSTNLIGS |
| 730 | 740 | 750 | 760 | 770 | 780 |
| GNFGNVFKGL | LGPENKLVAV | KVLNLLKHGA | TKSFMAECET | FKGIRHRNLV | KLITVCSSLD |
| 790 | 800 | 810 | 820 | 830 | 840 |
| SEGNDFRALV | YEFMPKGSLD | MWLQLEDLER | VNDHSRSLTP | AEKLNIAIDV | ASALEYLHVH |
| 850 | 860 | 870 | 880 | 890 | 900 |
| CHDPVAHCDI | KPSNILLDDD | LTAHVSDFGL | AQLLYKYDRE | SFLNQFSSAG | VRGTIGYAAP |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EYGMGGQPSI | QGDVYSFGIL | LLEMFSGKKP | TDESFAGDYN | LHSYTKSILS | GCTSSGGSNA |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| IDEGLRLVLQ | VGIKCSEEYP | RDRMRTDEAV | RELISIRSKF | FSSKTTITES | PRDAPQSSPQ |
| 1030 | |||||
| EWMLNTDMHT | M |