C0LGG3
Gene name |
At1g51820 (F19C24.23, T14L22.3) |
Protein name |
Probable LRR receptor-like serine/threonine-protein kinase At1g51820 |
Names |
|
Species |
Arabidopsis thaliana (Mouse-ear cress) |
KEGG Pathway |
ath:AT1G51820 |
EC number |
2.7.11.1: Protein-serine/threonine kinases |
Protein Class |
|
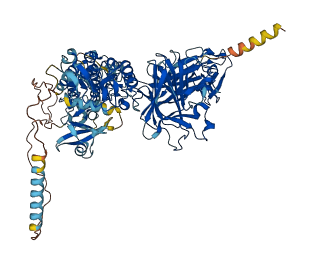
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
720-727 (Activation loop from InterPro)
Target domain |
578-851 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
738-745 (Activation loop from InterPro)
Target domain |
578-851 (Protein kinase domain) |
Relief mechanism |
|
Assay |
|
Autoinhibited structure

Activated structure

1 structures for C0LGG3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-C0LGG3-F1 | Predicted | AlphaFoldDB |
206 variants for C0LGG3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| ENSVATH04936882 | 21 | Q>* | No | 1000Genomes | |
| ENSVATH04936877 | 44 | Q>E | No | 1000Genomes | |
| tmp_1_19241652_T_A | 44 | Q>H | No | 1000Genomes | |
| ENSVATH04936876 | 50 | T>I | No | 1000Genomes | |
| ENSVATH04936876 | 50 | T>K | No | 1000Genomes | |
| ENSVATH14291028 | 50 | T>P | No | 1000Genomes | |
| tmp_1_19241624_C_T | 54 | D>N | No | 1000Genomes | |
| ENSVATH00093286 | 55 | L>F | No | 1000Genomes | |
| ENSVATH04936874 | 62 | G>S | No | 1000Genomes | |
| tmp_1_19241591_C_T | 65 | A>T | No | 1000Genomes | |
| ENSVATH04936873 | 66 | K>I | No | 1000Genomes | |
| tmp_1_19241515_T_A | 90 | Y>F | No | 1000Genomes | |
| tmp_1_19241505_A_T | 93 | N>K | No | 1000Genomes | |
| ENSVATH04936870 | 94 | V>F | No | 1000Genomes | |
| ENSVATH04936870 | 94 | V>I | No | 1000Genomes | |
| ENSVATH01347525 | 96 | S>R | No | 1000Genomes | |
| tmp_1_19241495_C_T | 97 | D>N | No | 1000Genomes | |
| ENSVATH01347524 | 101 | L>* | No | 1000Genomes | |
| tmp_1_19241470_G_A | 105 | T>I | No | 1000Genomes | |
| ENSVATH01347521 | 108 | Y>N | No | 1000Genomes | |
| tmp_1_19241459_C_T | 109 | G>R | No | 1000Genomes | |
| ENSVATH04936867 | 114 | L>F | No | 1000Genomes | |
| ENSVATH01347520 | 116 | V>I | No | 1000Genomes | |
| tmp_1_19241411_C_A | 125 | G>C | No | 1000Genomes | |
| tmp_1_19241371_A_T | 138 | I>K | No | 1000Genomes | |
| ENSVATH01347516 | 143 | L>H | No | 1000Genomes | |
| ENSVATH01347517 | 143 | L>I | No | 1000Genomes | |
| ENSVATH01347515 | 146 | R>K | No | 1000Genomes | |
| ENSVATH04936865 | 148 | N>I | No | 1000Genomes | |
| tmp_1_19241317_T_A | 156 | K>M | No | 1000Genomes | |
| tmp_1_19241314_G_A | 157 | T>M | No | 1000Genomes | |
| tmp_1_19241312_C_A | 158 | G>* | No | 1000Genomes | |
| tmp_1_19241293_A_T | 164 | I>K | No | 1000Genomes | |
| ENSVATH04936864 | 173 | K>M | No | 1000Genomes | |
| ENSVATH13511500 | 185 | K>T | No | 1000Genomes | |
| tmp_1_19241227_T_C | 186 | Y>C | No | 1000Genomes | |
| tmp_1_19241224_A_G | 187 | L>S | No | 1000Genomes | |
| ENSVATH04936863 | 190 | G>V | No | 1000Genomes | |
| ENSVATH04936862 | 195 | S>L | No | 1000Genomes | |
| ENSVATH13511499 | 197 | T>S | No | 1000Genomes | |
| tmp_1_19241188_A_T | 199 | I>K | No | 1000Genomes | |
| ENSVATH01347469 | 208 | R>H | No | 1000Genomes | |
| tmp_1_19240342_C_G | 210 | W>C | No | 1000Genomes | |
| ENSVATH04936831 | 215 | D>H | No | 1000Genomes | |
|
ENSVATH01347468 ENSVATH07644602 |
218 | W>* | No | 1000Genomes | |
| ENSVATH01347468 | 218 | W>L | No | 1000Genomes | |
| ENSVATH01347467 | 225 | L>F | No | 1000Genomes | |
| ENSVATH01347467 | 225 | L>I | No | 1000Genomes | |
| ENSVATH13511414 | 226 | K>* | No | 1000Genomes | |
| tmp_1_19240281_T_G | 231 | I>L | No | 1000Genomes | |
| ENSVATH01347465 | 242 | K>R | No | 1000Genomes | |
| ENSVATH04936830 | 245 | T>R | No | 1000Genomes | |
| ENSVATH04936829 | 246 | P>R | No | 1000Genomes | |
| ENSVATH01347464 | 256 | T>I | No | 1000Genomes | |
| ENSVATH04936828 | 265 | Q>* | No | 1000Genomes | |
| tmp_1_19240172_T_C | 267 | Y>C | No | 1000Genomes | |
| ENSVATH13511413 | 271 | H>D | No | 1000Genomes | |
| tmp_1_19240146_GA_CA,G | 276 | Q>E | No | 1000Genomes | |
| ENSVATH01347461 | 277 | A>V | No | 1000Genomes | |
| ENSVATH07644601 | 278 | L>I | No | 1000Genomes | |
| tmp_1_19240139_A_C | 278 | L>R | No | 1000Genomes | |
| ENSVATH07644601 | 278 | L>V | No | 1000Genomes | |
| tmp_1_19240133_G_A | 280 | A>V | No | 1000Genomes | |
| ENSVATH04936827 | 293 | E>A | No | 1000Genomes | |
| ENSVATH04936826 | 298 | P>T | No | 1000Genomes | |
| ENSVATH14290949 | 303 | P>A | No | 1000Genomes | |
| tmp_1_19240013_C_T | 320 | G>E | No | 1000Genomes | |
| tmp_1_19239990_T_A | 328 | K>* | No | 1000Genomes | |
| ENSVATH01347455 | 333 | T>R | No | 1000Genomes | |
| tmp_1_19239966_G_A | 336 | P>S | No | 1000Genomes | |
| ENSVATH01347451 | 337 | L>* | No | 1000Genomes | |
| tmp_1_19239960_G_C | 338 | L>V | No | 1000Genomes | |
| tmp_1_19239941_A_G | 344 | F>S | No | 1000Genomes | |
| ENSVATH13511410 | 345 | T>N | No | 1000Genomes | |
| ENSVATH01347445 | 356 | E>G | No | 1000Genomes | |
| ENSVATH01347444 | 357 | N>D | No | 1000Genomes | |
| ENSVATH01347444 | 357 | N>Y | No | 1000Genomes | |
| tmp_1_19239898_A_T | 358 | D>E | No | 1000Genomes | |
| ENSVATH13511409 | 360 | A>T | No | 1000Genomes | |
| ENSVATH01347434 | 367 | G>D | No | 1000Genomes | |
| ENSVATH01347432 | 371 | L>F | No | 1000Genomes | |
| ENSVATH01347433 | 371 | L>M | No | 1000Genomes | |
| tmp_1_19239748_A_T | 381 | C>S | No | 1000Genomes | |
| tmp_1_19239739_TG_AG,T | 384 | K>* | No | 1000Genomes | |
| ENSVATH13511408 | 385 | Q>L | No | 1000Genomes | |
| ENSVATH13511407 | 386 | L>F | No | 1000Genomes | |
| ENSVATH00093281 | 394 | K>N | No | 1000Genomes | |
| tmp_1_19239582_A_C | 409 | L>* | No | 1000Genomes | |
| ENSVATH04936817 | 419 | T>M | No | 1000Genomes | |
| ENSVATH14290946 | 433 | L>F | No | 1000Genomes | |
| tmp_1_19239366_C_T | 440 | G>E | No | 1000Genomes | |
| tmp_1_19239367_C_T | 440 | G>R | No | 1000Genomes | |
| tmp_1_19239364_C_T | 441 | E>K | No | 1000Genomes | |
| ENSVATH01347409 | 448 | D>E | No | 1000Genomes | |
| ENSVATH14290945 | 448 | D>N | No | 1000Genomes | |
| ENSVATH13511355 | 464 | G>D | No | 1000Genomes | |
| tmp_1_19239171_T_A | 474 | K>N | No | 1000Genomes | |
| ENSVATH04936813 | 475 | G>R | No | 1000Genomes | |
| ENSVATH01347398 | 476 | M>I | No | 1000Genomes | |
| ENSVATH13511354 | 479 | N>T | No | 1000Genomes | |
| ENSVATH01347394 | 480 | V>L | No | 1000Genomes | |
| ENSVATH04936810 | 481 | E>K | No | 1000Genomes | |
| ENSVATH04936809 | 482 | G>R | No | 1000Genomes | |
| ENSVATH01347392 | 487 | L>V | No | 1000Genomes | |
| tmp_1_19239045_T_C | 490 | T>A | No | 1000Genomes | |
| tmp_1_19239041_C_T | 491 | G>D | No | 1000Genomes | |
| ENSVATH00093280 | 492 | S>Y | No | 1000Genomes | |
| ENSVATH01347391 | 495 | K>N | No | 1000Genomes | |
| ENSVATH01347390 | 497 | K>R | No | 1000Genomes | |
| tmp_1_19239014_C_T | 500 | G>E | No | 1000Genomes | |
| ENSVATH13511344 | 513 | S>L | No | 1000Genomes | |
| ENSVATH13511343 | 518 | A>V | No | 1000Genomes | |
| ENSVATH01347388 | 521 | I>V | No | 1000Genomes | |
| tmp_1_19238942_A_G | 524 | L>S | No | 1000Genomes | |
| ENSVATH13511342 | 527 | F>L | No | 1000Genomes | |
| ENSVATH01347387 | 527 | F>S | No | 1000Genomes | |
| ENSVATH01347384 | 534 | R>K | No | 1000Genomes | |
| ENSVATH13511341 | 535 | S>P | No | 1000Genomes | |
| tmp_1_19238906_G_A | 536 | P>L | No | 1000Genomes | |
| ENSVATH04936803 | 552 | R>S | No | 1000Genomes | |
| ENSVATH13511335 | 557 | S>F | No | 1000Genomes | |
| ENSVATH04936802 | 564 | K>I | No | 1000Genomes | |
| tmp_1_19238716_T_A | 565 | N>Y | No | 1000Genomes | |
| ENSVATH13511322 | 574 | V>A | No | 1000Genomes | |
| ENSVATH13511320 | 580 | F>I | No | 1000Genomes | |
| tmp_1_19238626_C_T | 595 | G>S | No | 1000Genomes | |
| ENSVATH04936800 | 599 | G>S | No | 1000Genomes | |
| tmp_1_19238591_C_G | 606 | K>N | No | 1000Genomes | |
| ENSVATH01347379 | 610 | H>D | No | 1000Genomes | |
| ENSVATH01347377 | 618 | Q>E | No | 1000Genomes | |
| ENSVATH04936799 | 618 | Q>L | No | 1000Genomes | |
| ENSVATH01347363 | 624 | E>D | No | 1000Genomes | |
| ENSVATH01347362 | 625 | L>I | No | 1000Genomes | |
| tmp_1_19238436_G_C | 627 | L>V | No | 1000Genomes | |
| tmp_1_19238426_T_G | 630 | H>P | No | 1000Genomes | |
| ENSVATH04936796 | 631 | H>D | No | 1000Genomes | |
| ENSVATH14290916 | 636 | G>R | No | 1000Genomes | |
| ENSVATH01347357 | 641 | C>* | No | 1000Genomes | |
| tmp_1_19238393_C_A | 641 | C>F | No | 1000Genomes | |
| tmp_1_19238391_C_T | 642 | D>N | No | 1000Genomes | |
| ENSVATH01347355 | 643 | E>G | No | 1000Genomes | |
| ENSVATH01347356 | 643 | E>Q | No | 1000Genomes | |
| ENSVATH13511316 | 644 | G>E | No | 1000Genomes | |
| ENSVATH01347354 | 645 | D>E | No | 1000Genomes | |
| tmp_1_19238382_C_T | 645 | D>N | No | 1000Genomes | |
| ENSVATH13511315 | 647 | L>M | No | 1000Genomes | |
| ENSVATH01347353 | 649 | L>F | No | 1000Genomes | |
| ENSVATH01347352 | 663 | M>L | No | 1000Genomes | |
| tmp_1_19238324_G_A | 664 | S>L | No | 1000Genomes | |
| ENSVATH01347345 | 668 | N>K | No | 1000Genomes | |
| tmp_1_19238222_C_A | 669 | R>L | No | 1000Genomes | |
| tmp_1_19238211_T_G | 673 | N>H | No | 1000Genomes | |
| ENSVATH04936790 | 685 | A>G | No | 1000Genomes | |
| ENSVATH04936789 | 686 | Q>K | No | 1000Genomes | |
| ENSVATH04936787 | 695 | C>Y | No | 1000Genomes | |
| ENSVATH04936784 | 709 | I>L | No | 1000Genomes | |
| ENSVATH14290893 | 711 | L>F | No | 1000Genomes | |
| ENSVATH14290892 | 717 | A>T | No | 1000Genomes | |
| ENSVATH04936783 | 724 | L>F | No | 1000Genomes | |
| tmp_1_19237974_C_T | 726 | R>K | No | 1000Genomes | |
| ENSVATH01347324 | 729 | L>P | No | 1000Genomes | |
| tmp_1_19237963_T_C | 730 | I>V | No | 1000Genomes | |
| ENSVATH04936782 | 735 | H>Y | No | 1000Genomes | |
| ENSVATH04936781 | 736 | V>G | No | 1000Genomes | |
| tmp_1_19237935_A_G | 739 | V>A | No | 1000Genomes | |
| tmp_1_19237933_C_T | 740 | V>I | No | 1000Genomes | |
| ENSVATH01347319 | 742 | G>E | No | 1000Genomes | |
| ENSVATH13511272 | 745 | G>R | No | 1000Genomes | |
| ENSVATH04936779 | 747 | L>I | No | 1000Genomes | |
| ENSVATH04936778 | 750 | E>G | No | 1000Genomes | |
| ENSVATH01347313 | 752 | H>Y | No | 1000Genomes | |
| tmp_1_19237807_C_T | 753 | R>K | No | 1000Genomes | |
| ENSVATH04936777 | 765 | S>N | No | 1000Genomes | |
| ENSVATH01347308 | 769 | L>V | No | 1000Genomes | |
| ENSVATH01347306 | 773 | I>L | No | 1000Genomes | |
| ENSVATH01347305 | 775 | T>I | No | 1000Genomes | |
| tmp_1_19237736_G_C | 777 | R>G | No | 1000Genomes | |
| ENSVATH01347303 | 778 | H>L | No | 1000Genomes | |
| ENSVATH01347301 | 784 | R>H | No | 1000Genomes | |
| ENSVATH01347299 | 790 | G>A | No | 1000Genomes | |
| ENSVATH04936776 | 791 | E>K | No | 1000Genomes | |
| ENSVATH01347297 | 795 | V>E | No | 1000Genomes | |
| ENSVATH01347298 | 795 | V>L | No | 1000Genomes | |
| ENSVATH01347295 | 803 | Q>H | No | 1000Genomes | |
| ENSVATH01347296 | 803 | Q>K | No | 1000Genomes | |
| tmp_1_19237639_C_T | 809 | S>N | No | 1000Genomes | |
| ENSVATH07644577 | 810 | L>F | No | 1000Genomes | |
| ENSVATH07644576 | 813 | D>N | No | 1000Genomes | |
| ENSVATH07644575 | 817 | G>C | No | 1000Genomes | |
| tmp_1_19237606_C_T | 820 | W>* | No | 1000Genomes | |
| tmp_1_19237588_G_A | 826 | A>V | No | 1000Genomes | |
| ENSVATH14290890 | 828 | S>N | No | 1000Genomes | |
| ENSVATH01347290 | 832 | H>P | No | 1000Genomes | |
| tmp_1_19237545_C_G | 840 | M>I | No | 1000Genomes | |
| ENSVATH04936770 | 850 | C>* | No | 1000Genomes | |
| tmp_1_19237505_C_T | 854 | E>K | No | 1000Genomes | |
| tmp_1_19237498_G_A | 856 | A>V | No | 1000Genomes | |
| ENSVATH04936769 | 864 | M>I | No | 1000Genomes | |
| tmp_1_19237464_C_A | 867 | K>N | No | 1000Genomes | |
| tmp_1_19237462_C_T | 868 | S>N | No | 1000Genomes | |
| ENSVATH07644564 | 877 | G>D | No | 1000Genomes | |
| tmp_1_19237436_C_T | 877 | G>S | No | 1000Genomes | |
| ENSVATH00093279 | 882 | P>R | No | 1000Genomes | |
| tmp_1_19237416_G_C | 883 | N>K | No | 1000Genomes | |
| tmp_1_19237417_T_G | 883 | N>T | No | 1000Genomes | |
| ENSVATH04936765 | 885 | R>Q | No | 1000Genomes |
No associated diseases with C0LGG3
5 regional properties for C0LGG3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Protein kinase domain | 578 - 851 | IPR000719 |
| repeat | Leucine-rich repeat | 405 - 462 | IPR001611 |
| active_site | Serine/threonine-protein kinase, active site | 699 - 711 | IPR008271 |
| binding_site | Protein kinase, ATP binding site | 584 - 606 | IPR017441 |
| domain | Malectin-like domain | 29 - 346 | IPR024788 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.11.1 | Protein-serine/threonine kinases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| integral component of membrane | The component of a membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| protein serine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate. |
| protein serine/threonine kinase activity | Catalysis of the reactions: ATP + protein serine = ADP + protein serine phosphate, and ATP + protein threonine = ADP + protein threonine phosphate. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| protein phosphorylation | The process of introducing a phosphate group on to a protein. |
28 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P20594 | NPR2 | Atrial natriuretic peptide receptor 2 | Homo sapiens (Human) | PR |
| Q03145 | Epha2 | Ephrin type-A receptor 2 | Mus musculus (Mouse) | PR |
| Q60750 | Epha1 | Ephrin type-A receptor 1 | Mus musculus (Mouse) | SS |
| Q924U5 | Tesk2 | Dual specificity testis-specific protein kinase 2 | Rattus norvegicus (Rat) | PR |
| P43293 | PBL11 | Probable serine/threonine-protein kinase PBL11 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q3E8W4 | ANX2 | Receptor-like protein kinase ANXUR2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8LPS5 | SERK5 | Somatic embryogenesis receptor kinase 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGW2 | PAM74 | Probable LRR receptor-like serine/threonine-protein kinase PAM74 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN93 | At5g59680 | Probable LRR receptor-like serine/threonine-protein kinase At5g59680 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9FN94 | At5g59670 | Receptor-like protein kinase At5g59670 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LIG2 | At3g21340 | Receptor-like protein kinase At3g21340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SNA3 | At3g46340 | Putative receptor-like protein kinase At3g46340 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9ZQR3 | At2g14510 | Leucine-rich repeat receptor-like serine/threonine-protein kinase At2g14510 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LPZ9 | SD113 | G-type lectin S-receptor-like serine/threonine-protein kinase SD1-13 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| P43298 | TMK1 | Receptor protein kinase TMK1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| C0LGI2 | At1g67720 | Probable LRR receptor-like serine/threonine-protein kinase At1g67720 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWW0 | ALE2 | Receptor-like serine/threonine-protein kinase ALE2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| P93050 | BSH | Probable LRR receptor-like serine/threonine-protein kinase RKF3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9LRP3 | At3g17420 | Probable receptor-like protein kinase At3g17420 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M2S4 | LECRKS4 | L-type lectin-domain containing receptor kinase S.4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8RWZ5 | SD25 | G-type lectin S-receptor-like serine/threonine-protein kinase SD2-5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64556 | At2g19230 | Putative leucine-rich repeat receptor-like serine/threonine-protein kinase At2g19230 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9SXB4 | At1g11300 | G-type lectin S-receptor-like serine/threonine-protein kinase At1g11300 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q84JQ4 | PRK2 | Pollen receptor-like kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q93ZS4 | NIK3 | Protein NSP-INTERACTING KINASE 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q94AG2 | SERK1 | Somatic embryogenesis receptor kinase 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| O64477 | At2g19130 | G-type lectin S-receptor-like serine/threonine-protein kinase At2g19130 | Arabidopsis thaliana (Mouse-ear cress) | SS |
| Q9XIC7 | SERK2 | Somatic embryogenesis receptor kinase 2 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MERHFVFIAT | YLLIFHLVQA | QNQTGFISVD | CGLSLLESPY | DAPQTGLTYT | SDADLVASGK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| TGRLAKEFEP | LVDKPTLTLR | YFPEGVRNCY | NLNVTSDTNY | LIKATFVYGN | YDGLNVGPNF |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NLYLGPNLWT | TVSSNDTIEE | IILVTRSNSL | QVCLVKTGIS | IPFINMLELR | PMKKNMYVTQ |
| 190 | 200 | 210 | 220 | 230 | 240 |
| SGSLKYLFRG | YISNSSTRIR | FPDDVYDRKW | YPLFDDSWTQ | VTTNLKVNTS | ITYELPQSVM |
| 250 | 260 | 270 | 280 | 290 | 300 |
| AKAATPIKAN | DTLNITWTVE | PPTTQFYSYV | HIAEIQALRA | NETREFNVTL | NGEYTFGPFS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PIPLKTASIV | DLSPGQCDGG | RCILQVVKTL | KSTLPPLLNA | IEAFTVIDFP | QMETNENDVA |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GIKNVQGTYG | LSRISWQGDP | CVPKQLLWDG | LNCKNSDIST | PPIITSLDLS | SSGLTGIITQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AIKNLTHLQI | LDLSDNNLTG | EVPEFLADIK | SLLVINLSGN | NLSGSVPPSL | LQKKGMKLNV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| EGNPHILCTT | GSCVKKKEDG | HKKKSVIVPV | VASIASIAVL | IGALVLFLIL | RKKRSPKVEG |
| 550 | 560 | 570 | 580 | 590 | 600 |
| PPPSYMQASD | GRLPRSSEPA | IVTKNRRFSY | SQVVIMTNNF | QRILGKGGFG | MVYHGFVNGT |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EQVAVKILSH | SSSQGYKQFK | AEVELLLRVH | HKNLVGLVGY | CDEGDNLALI | YEYMANGDLK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| EHMSGTRNRF | ILNWGTRLKI | VIESAQGLEY | LHNGCKPPMV | HRDVKTTNIL | LNEHFEAKLA |
| 730 | 740 | 750 | 760 | 770 | 780 |
| DFGLSRSFLI | EGETHVSTVV | AGTPGYLDPE | YHRTNWLTEK | SDVYSFGILL | LEIITNRHVI |
| 790 | 800 | 810 | 820 | 830 | 840 |
| DQSREKPHIG | EWVGVMLTKG | DIQSIMDPSL | NEDYDSGSVW | KAVELAMSCL | NHSSARRPTM |
| 850 | 860 | 870 | 880 | ||
| SQVVIELNEC | LASENARGGA | SRDMESKSSI | EVSLTFGTEV | SPNAR |