B2RX12
Gene name |
Abcc3 (Cmoat2, Mrp3) |
Protein name |
ATP-binding cassette sub-family C member 3 |
Names |
Canalicular multispecific organic anion transporter 2, Multidrug resistance-associated protein 3 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:76408 |
EC number |
7.6.2.3: Linked to the hydrolysis of a nucleoside triphosphate |
Protein Class |
|
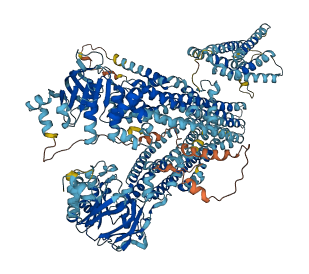
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for B2RX12
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-B2RX12-F1 | Predicted | AlphaFoldDB |
86 variants for B2RX12
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3389191614 | 2 | D>N | No | EVA | |
| rs3389168087 | 10 | L>Q | No | EVA | |
| rs27076245 | 22 | Y>H | No | EVA | |
| rs3389207678 | 41 | P>L | No | EVA | |
| rs3389128361 | 80 | L>P | No | EVA | |
| rs3389192845 | 101 | P>H | No | EVA | |
| rs3389155816 | 112 | V>M | No | EVA | |
| rs3389181064 | 119 | A>V | No | EVA | |
| rs3389196802 | 128 | L>F | No | EVA | |
| rs3389192723 | 131 | V>I | No | EVA | |
| rs212780065 | 159 | A>V | No | EVA | |
| rs3389186845 | 230 | G>S | No | EVA | |
| rs3389196742 | 243 | L>V | No | EVA | |
| rs13481166 | 269 | R>G | No | EVA | |
| rs16790889 | 274 | T>P | No | EVA | |
| rs16790890 | 309 | S>N | No | EVA | |
| rs16790891 | 317 | N>K | No | EVA | |
| rs16790892 | 321 | N>D | No | EVA | |
| rs3389193147 | 351 | L>M | No | EVA | |
| rs16783395 | 357 | L>M | No | EVA | |
| rs16783394 | 365 | I>M | No | EVA | |
| rs3402243277 | 371 | H>Y | No | EVA | |
| rs16783393 | 426 | I>L | No | EVA | |
| rs3389201090 | 437 | I>V | No | EVA | |
| rs3402391158 | 472 | M>* | No | EVA | |
| rs3389168135 | 506 | E>K | No | EVA | |
| rs234206305 | 558 | E>K | No | EVA | |
| rs3389155812 | 560 | N>S | No | EVA | |
| rs3389207627 | 583 | M>I | No | EVA | |
| rs3389128399 | 621 | S>F | No | EVA | |
| rs3389181069 | 632 | T>I | No | EVA | |
| rs3389190495 | 659 | V>M | No | EVA | |
| rs3389186887 | 659 | V>S | No | EVA | |
| rs3389191609 | 665 | G>R | No | EVA | |
| rs3389181059 | 693 | V>M | No | EVA | |
| rs3389196793 | 703 | T>I | No | EVA | |
| rs3389186878 | 750 | L>I | No | EVA | |
| rs3389162957 | 766 | S>N | No | EVA | |
| rs3389201152 | 784 | V>E | No | EVA | |
| rs3389196766 | 791 | Q>* | No | EVA | |
| rs3389162940 | 814 | L>M | No | EVA | |
| rs3389162967 | 825 | G>D | No | EVA | |
| rs27076267 | 829 | S>A | No | EVA | |
| rs3389203662 | 830 | E>D | No | EVA | |
| rs3389192803 | 833 | H>R | No | EVA | |
| rs3389193158 | 838 | L>P | No | EVA | |
| rs243526961 | 843 | S>P | No | EVA | |
| rs3389190514 | 846 | N>K | No | EVA | |
| rs221592969 | 894 | V>I | No | EVA | |
| rs3389155808 | 910 | E>K | No | EVA | |
| rs3389201100 | 914 | R>L | No | EVA | |
| rs49731813 | 919 | K>R | No | EVA | |
| rs3389181073 | 922 | N>Y | No | EVA | |
| rs3412846250 | 928 | A>V | No | EVA | |
| rs3389193186 | 933 | T>I | No | EVA | |
| rs3389168082 | 936 | T>I | No | EVA | |
| rs3389181042 | 955 | Y>H | No | EVA | |
| rs261794182 | 968 | L>I | No | EVA | |
| rs3389203668 | 990 | A>T | No | EVA | |
| rs3402638865 | 995 | A>V | No | EVA | |
| rs3389186843 | 1060 | P>A | No | EVA | |
| rs211947932 | 1067 | R>C | No | EVA | |
| rs3402519881 | 1078 | V>A | No | EVA | |
| rs3389193184 | 1153 | G>D | No | EVA | |
| rs3389191621 | 1174 | V>M | No | EVA | |
| rs3389186894 | 1189 | R>* | No | EVA | |
| rs3401980827 | 1228 | Y>H | No | EVA | |
| rs3401980835 | 1228 | Y>S | No | EVA | |
| rs3402702541 | 1231 | Q>H | No | EVA | |
| rs3402802901 | 1231 | Q>P | No | EVA | |
| rs3389128371 | 1239 | M>V | No | EVA | |
| rs29473877 | 1263 | K>E | No | EVA | |
| rs234540646 | 1307 | V>I | No | EVA | |
| rs3389203660 | 1317 | I>V | No | EVA | |
| rs248321069 | 1344 | V>F | No | EVA | |
| rs238863988 | 1401 | N>S | No | EVA | |
| rs3402014921 | 1446 | D>N | No | EVA | |
| rs3402728358 | 1447 | E>Q | No | EVA | |
| rs3389196804 | 1468 | F>L | No | EVA | |
| rs3402939074 | 1480 | L>I | No | EVA | |
| rs3402638902 | 1481 | N>Y | No | EVA | |
| rs3389128396 | 1489 | V>I | No | EVA | |
| rs3389196219 | 1507 | I>M | No | EVA | |
| rs3389193183 | 1511 | G>S | No | EVA | |
| rs3389192794 | 1522 | L>F | No | EVA | |
| rs3389207663 | 1523 | A>S | No | EVA |
No associated diseases with B2RX12
8 regional properties for B2RX12
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | ABC transporter-like, ATP-binding domain | 626 - 850 | IPR003439-1 |
| domain | ABC transporter-like, ATP-binding domain | 1287 - 1519 | IPR003439-2 |
| domain | AAA+ ATPase domain | 652 - 827 | IPR003593-1 |
| domain | AAA+ ATPase domain | 1311 - 1496 | IPR003593-2 |
| domain | ABC transporter type 1, transmembrane domain | 311 - 593 | IPR011527-1 |
| domain | ABC transporter type 1, transmembrane domain | 970 - 1248 | IPR011527-2 |
| conserved_site | ABC transporter-like, conserved site | 750 - 764 | IPR017871-1 |
| conserved_site | ABC transporter-like, conserved site | 1422 - 1436 | IPR017871-2 |
Functions
| Description | ||
|---|---|---|
| EC Number | 7.6.2.3 | Linked to the hydrolysis of a nucleoside triphosphate |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| basolateral plasma membrane | The region of the plasma membrane that includes the basal end and sides of the cell. Often used in reference to animal polarized epithelial membranes, where the basal membrane is the part attached to the extracellular matrix, or in plant cells, where the basal membrane is defined with respect to the zygotic axis. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
8 GO annotations of molecular function
| Name | Definition |
|---|---|
| ABC-type bile acid transporter activity | Enables the transfer of a solute or solutes from one side of a membrane to the other according to the reaction: bile acid(in) + ATP + H2O -> bile acid(out) + ADP + phosphate. |
| ABC-type glutathione S-conjugate transporter activity | Catalysis of the reaction: ATP + H2O + glutathione S-conjugate(in) -> ADP + phosphate + glutathione S-conjugate(out). |
| ABC-type xenobiotic transporter activity | Catalysis of the reaction: ATP + H2O + xenobiotic(in) = ADP + phosphate + xenobiotic(out). |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| ATPase-coupled transmembrane transporter activity | Primary active transporter of a solute across a membrane, via the reaction: ATP + H2O = ADP + phosphate, to directly drive the transport of a substance across a membrane. The transport protein may be transiently phosphorylated (P-type transporters), or not (ABC-type transporters and other families of transporters). Primary active transport occurs up the solute's concentration gradient and is driven by a primary energy source. |
| glucuronoside transmembrane transporter activity | Enables the transfer of a glucuronosides from one side of a membrane to the other. Glucuronosides are any compound formed by combination of glycosidic linkage of a hydroxy compound (e.g. an alcohol or a saccharide) with the anomeric carbon atom of glucuronate. |
| icosanoid transmembrane transporter activity | Enables the transfer of icosanoids from one side of a membrane to the other. |
| xenobiotic transmembrane transporter activity | Enables the directed movement of a xenobiotic from one side of a membrane to the other. A xenobiotic is a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
7 GO annotations of biological process
| Name | Definition |
|---|---|
| anion transmembrane transport | The process in which an anion is transported across a membrane. |
| bile acid and bile salt transport | The directed movement of bile acid and bile salts into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. |
| canalicular bile acid transport | Enables the transfer of bile acid from one side of a hepatocyte plasma membrane into a bile canaliculus. Bile canaliculi are the thin tubes formed by hepatocyte membranes. Bile acids are any of a group of steroid carboxylic acids occurring in bile, where they are present as the sodium salts of their amides with glycine or taurine. |
| leukotriene transport | The directed movement of leukotrienes into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. Leukotrienes are linear C20 endogenous metabolites of arachidonic acid (icosa-5,8,11,14-tetraenoic acid) containing a terminal carboxy function and four or more double bonds (three or more of which are conjugated) as well as other functional groups. |
| transmembrane transport | The process in which a solute is transported across a lipid bilayer, from one side of a membrane to the other. |
| xenobiotic transmembrane transport | The process in which a xenobiotic, a compound foreign to the organim exposed to it, is transported across a membrane. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
| xenobiotic transport | The directed movement of a xenobiotic into, out of or within a cell, or between cells, by means of some agent such as a transporter or pore. A xenobiotic is a compound foreign to the organim exposed to it. It may be synthesized by another organism (like ampicilin) or it can be a synthetic chemical. |
25 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P14772 | BPT1 | Bile pigment transporter 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q8HXQ5 | ABCC1 | Multidrug resistance-associated protein 1 | Bos taurus (Bovine) | PR |
| Q2QLE5 | CFTR | Cystic fibrosis transmembrane conductance regulator | Pan troglodytes (Chimpanzee) | PR |
| P91660 | Rh5 | Probable multidrug resistance-associated protein lethal(2)03659 | Drosophila melanogaster (Fruit fly) | PR |
| P13569 | CFTR | Cystic fibrosis transmembrane conductance regulator | Homo sapiens (Human) | PR |
| O95255 | ABCC6 | ATP-binding cassette sub-family C member 6 | Homo sapiens (Human) | PR |
| Q92887 | ABCC2 | ATP-binding cassette sub-family C member 2 | Homo sapiens (Human) | SS |
| O15439 | ABCC4 | ATP-binding cassette sub-family C member 4 | Homo sapiens (Human) | PR |
| Q96J66 | ABCC11 | ATP-binding cassette sub-family C member 11 | Homo sapiens (Human) | PR |
| P33527 | ABCC1 | Multidrug resistance-associated protein 1 | Homo sapiens (Human) | PR |
| O15438 | ABCC3 | ATP-binding cassette sub-family C member 3 | Homo sapiens (Human) | PR |
| O35379 | Abcc1 | Multidrug resistance-associated protein 1 | Mus musculus (Mouse) | PR |
| Q8VI47 | Abcc2 | ATP-binding cassette sub-family C member 2 | Mus musculus (Mouse) | SS |
| Q80WJ6 | Abcc12 | ATP-binding cassette sub-family C member 12 | Mus musculus (Mouse) | PR |
| P26361 | Cftr | Cystic fibrosis transmembrane conductance regulator | Mus musculus (Mouse) | PR |
| Q6PQZ2 | CFTR | Cystic fibrosis transmembrane conductance regulator | Sus scrofa (Pig) | PR |
| Q6Y306 | Abcc12 | ATP-binding cassette sub-family C member 12 | Rattus norvegicus (Rat) | PR |
| Q8CG09 | Abcc1 | Multidrug resistance-associated protein 1 | Rattus norvegicus (Rat) | PR |
| Q63120 | Abcc2 | ATP-binding cassette sub-family C member 2 | Rattus norvegicus (Rat) | EV |
| Q00553 | CFTR | Cystic fibrosis transmembrane conductance regulator | Macaca mulatta (Rhesus macaque) | PR |
| Q9SKX0 | ABCC13 | ABC transporter C family member 13 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q8VZZ4 | ABCC6 | ABC transporter C family member 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9M1C7 | ABCC9 | ABC transporter C family member 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C8G9 | ABCC1 | ABC transporter C family member 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9C8H0 | ABCC12 | ABC transporter C family member 12 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MDRLCGSGEL | GSKFWDSNLS | IYTNTPDLTP | CFQNSLLAWV | PCIYLWAALP | CYLFYLRHHQ |
| 70 | 80 | 90 | 100 | 110 | 120 |
| LGYIVLSWLS | RLKTALGVLL | WCVSWVDLFY | SFHGLIHGSS | PAPVFFVTPL | VVGITMLLAT |
| 130 | 140 | 150 | 160 | 170 | 180 |
| LLIQYERLRG | VQSSGVLIIF | WLLCVICAII | PFRSKILSAL | AEGKILDPFR | FTTFYIYFAL |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VFCALILSCF | KEKPPLFSPE | NLDTNPCPEA | SAGFFSRLSF | WWFTRLAILG | YRRPLEDRDL |
| 250 | 260 | 270 | 280 | 290 | 300 |
| WSLSEEDCSH | KVVQRLLEAW | QKQQNQASRS | QTATAEPKIP | GEDAVLLKPR | PKSKQPSFLR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| ALVRTFTSSL | LMSACFNLIQ | NLLGFVNPQL | LSILIRFISD | PTAPTWWGFL | LAGLMFLSST |
| 370 | 380 | 390 | 400 | 410 | 420 |
| MQTLILHQYY | HCIFVMALRL | RTAIIGVIYR | KALVITNSVK | RESTVGEMVN | LMSVDAQRFM |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DVSPFINLLW | SAPLQVILAI | YFLWQILGPS | ALAGVAVIVL | LIPLNGAVSM | KMKTYQVKQM |
| 490 | 500 | 510 | 520 | 530 | 540 |
| KFKDSRIKLM | SEILNGIKVL | KLYAWEPSFL | EQVKGIRQSE | LQLLRKGAYL | QAISTFIWIC |
| 550 | 560 | 570 | 580 | 590 | 600 |
| TPFLVTLITL | GVYVYVDESN | VLDAEKAFVS | LSLFNILKIP | LNMLPQLISG | LTQASVSLKR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| IQDFLNQNEL | DPQCVERKTI | SPGYAITIHN | GTFTWAQDLP | PTLHSLNIQI | PKGALVAVVG |
| 670 | 680 | 690 | 700 | 710 | 720 |
| PVGCGKSSLV | SALLGEMEKL | EGVVSVKGSV | AYVPQQAWIQ | NCTLQENVLF | GQPMNPKRYQ |
| 730 | 740 | 750 | 760 | 770 | 780 |
| QALETCALLA | DLDVLPGGDQ | TEIGEKGINL | SGGQRQRVSL | ARAVYSDANI | FLLDDPLSAV |
| 790 | 800 | 810 | 820 | 830 | 840 |
| DSHVAKHIFD | QVIGPEGVLA | GKTRVLVTHG | ISFLPQTDFI | IVLAGGQVSE | MGHYSALLQH |
| 850 | 860 | 870 | 880 | 890 | 900 |
| DGSFANFLRN | YAPDEDQEDH | EAALQNANEE | VLLLEDTLST | HTDLTDNEPA | IYEVRKQFMR |
| 910 | 920 | 930 | 940 | 950 | 960 |
| EMSSLSSEGE | VQNRTMPKKH | TNSLEKEALV | TKTKETGALI | KEEIAETGNV | KLSVYWDYAK |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| SMGLCTTLSI | CLLYGGQSAA | AIGANVWLSA | WSNDAEEHGQ | QNKTSVRLGV | YAALGILQGL |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| LVMLSAFTMV | VGAIQAARLL | HEALLHNKIR | SPQSFFDTTP | SGRILNRFSK | DIYVIDEVLA |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| PTILMLLNSF | FTSISTIMVI | VASTPLFMVV | VLPLAVLYGF | VQRFYVATSR | QLKRLESISR |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| SPIFSHFSET | VTGTSVIRAY | GRIQDFKVLS | DTKVDNNQKS | SYPYIASNRW | LGVHVEFVGN |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| CVVLFAALFA | VIGRNSLNPG | LVGLSVSYAL | QVTMALNWMI | RMISDLESNI | IAVERVKEYS |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| KTKTEAPWVV | ESNRAPEGWP | TRGMVEFRNY | SVRYRPGLEL | VLKNVTVHVQ | GGEKVGIVGR |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| TGAGKSSMTL | CLFRILEAAE | GEIVIDGLNV | AHIGLHDLRS | QLTIIPQDPI | LFSGTLRMNL |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| DPFGRYSEED | IWRALELSHL | NTFVSSQPAG | LDFQCAEGGD | NLSVGQRQLV | CLARALLRKS |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| RVLVLDEATA | AIDLETDDLI | QGTIRTQFED | CTVLTIAHRL | NTIMDYNRVL | VLDKGVVAEF |
| 1510 | 1520 | ||||
| DSPVNLIAAG | GIFYGMAKDA | GLA |