A9JRZ0
Gene name |
smurf2 |
Protein name |
E3 ubiquitin-protein ligase SMURF2 |
Names |
EC 2.3.2.26 , HECT-type E3 ubiquitin transferase SMURF2 , SMAD ubiquitination regulatory factor 2 , SMAD-specific E3 ubiquitin-protein ligase 2 |
Species |
Danio rerio (Zebrafish) (Brachydanio rerio) |
KEGG Pathway |
dre:563633 |
EC number |
2.3.2.26: Aminoacyltransferases |
Protein Class |
|
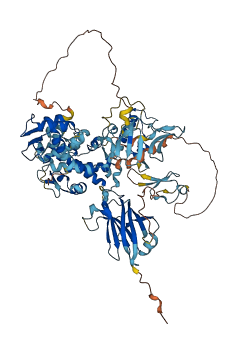
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
410-765 (HECT domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
References
- Yeon JH et al. (2016) "Systems-wide Identification of cis-Regulatory Elements in Proteins", Cell systems, 2, 89-100
- Wiesner S et al. (2007) "Autoinhibition of the HECT-type ubiquitin ligase Smurf2 through its C2 domain", Cell, 130, 651-62
- Lu K et al. (2011) "Pivotal role of the C2 domain of the Smurf1 ubiquitin ligase in substrate selection", The Journal of biological chemistry, 286, 16861-70
- Ogunjimi AA et al. (2005) "Regulation of Smurf2 ubiquitin ligase activity by anchoring the E2 to the HECT domain", Molecular cell, 19, 297-308
Autoinhibited structure

Activated structure

1 structures for A9JRZ0
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A9JRZ0-F1 | Predicted | AlphaFoldDB |
No variants for A9JRZ0
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for A9JRZ0 | |||||
No associated diseases with A9JRZ0
5 regional properties for A9JRZ0
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 1 - 117 | IPR000008 |
| domain | HECT domain | 410 - 765 | IPR000569 |
| domain | WW domain | 157 - 190 | IPR001202-1 |
| domain | WW domain | 251 - 284 | IPR001202-2 |
| domain | WW domain | 297 - 330 | IPR001202-3 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.3.2.26 | Aminoacyltransferases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| membrane raft | Any of the small (10-200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched membrane domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| SMAD binding | Binding to a SMAD signaling protein. |
| ubiquitin protein ligase activity | Catalysis of the transfer of ubiquitin to a substrate protein via the reaction X-ubiquitin + S -> X + S-ubiquitin, where X is either an E2 or E3 enzyme, the X-ubiquitin linkage is a thioester bond, and the S-ubiquitin linkage is an amide bond |
4 GO annotations of biological process
| Name | Definition |
|---|---|
| negative regulation of BMP signaling pathway | Any process that stops, prevents, or reduces the frequency, rate or extent of the BMP signaling pathway. |
| proteasome-mediated ubiquitin-dependent protein catabolic process | The chemical reactions and pathways resulting in the breakdown of a protein or peptide by hydrolysis of its peptide bonds, initiated by the covalent attachment of ubiquitin, and mediated by the proteasome. |
| protein polyubiquitination | Addition of multiple ubiquitin groups to a protein, forming a ubiquitin chain. |
| protein ubiquitination | The process in which one or more ubiquitin groups are added to a protein. |
23 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P39940 | RSP5 | E3 ubiquitin-protein ligase RSP5 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| Q9Y0H4 | Su(dx) | E3 ubiquitin-protein ligase Su | Drosophila melanogaster (Fruit fly) | SS |
| Q9V853 | Smurf | E3 ubiquitin-protein ligase Smurf1 | Drosophila melanogaster (Fruit fly) | SS |
| O00308 | WWP2 | NEDD4-like E3 ubiquitin-protein ligase WWP2 | Homo sapiens (Human) | EV |
| Q9HCE7 | SMURF1 | E3 ubiquitin-protein ligase SMURF1 | Homo sapiens (Human) | PR |
| O95817 | BAG3 | BAG family molecular chaperone regulator 3 | Homo sapiens (Human) | PR |
| Q96PU5 | NEDD4L | E3 ubiquitin-protein ligase NEDD4-like | Homo sapiens (Human) | PR |
| O60861 | GAS7 | Growth arrest-specific protein 7 | Homo sapiens (Human) | PR |
| P46934 | NEDD4 | E3 ubiquitin-protein ligase NEDD4 | Homo sapiens (Human) | EV |
| Q9H0M0 | WWP1 | NEDD4-like E3 ubiquitin-protein ligase WWP1 | Homo sapiens (Human) | EV |
| Q96J02 | ITCH | E3 ubiquitin-protein ligase Itchy homolog | Homo sapiens (Human) | EV |
| Q9HAU4 | SMURF2 | E3 ubiquitin-protein ligase SMURF2 | Homo sapiens (Human) | EV |
| P46935 | Nedd4 | E3 ubiquitin-protein ligase NEDD4 | Mus musculus (Mouse) | PR |
| Q9CUN6 | Smurf1 | E3 ubiquitin-protein ligase SMURF1 | Mus musculus (Mouse) | PR |
| Q8BZZ3 | Wwp1 | NEDD4-like E3 ubiquitin-protein ligase WWP1 | Mus musculus (Mouse) | SS |
| Q60780 | Gas7 | Growth arrest-specific protein 7 | Mus musculus (Mouse) | PR |
| Q8C863 | Itch | E3 ubiquitin-protein ligase Itchy | Mus musculus (Mouse) | EV |
| Q9DBH0 | Wwp2 | NEDD4-like E3 ubiquitin-protein ligase WWP2 | Mus musculus (Mouse) | SS |
| Q8CFI0 | Nedd4l | E3 ubiquitin-protein ligase NEDD4-like | Mus musculus (Mouse) | PR |
| A2A5Z6 | Smurf2 | E3 ubiquitin-protein ligase SMURF2 | Mus musculus (Mouse) | SS |
| Q62940 | Nedd4 | E3 ubiquitin-protein ligase NEDD4 | Rattus norvegicus (Rat) | PR |
| Q9N2Z7 | wwp-1 | E3 ubiquitin-protein ligase wwp-1 | Caenorhabditis elegans | SS |
| F8W2M1 | hace1 | E3 ubiquitin-protein ligase HACE1 | Danio rerio (Zebrafish) (Brachydanio rerio) | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSNQGVRRNG | PVKLRLTVLC | AKNLVKKDFF | RLPDPFAKVV | VDGSGQCHST | DTVRNTLDPK |
| 70 | 80 | 90 | 100 | 110 | 120 |
| WNQHYDLYIG | KSDSITISVW | NHKKIHKKQG | AGFLGCVRLL | SNSINRLKDT | GYQRLDLNKL |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GPNDSDTVRG | QIVVSLQSRD | RIGSGGPVVD | CSRLFDNDLP | DGWEERRTAS | GRIQYLNHIT |
| 190 | 200 | 210 | 220 | 230 | 240 |
| RSTQWERPTR | PASEYSSPGR | PLSCLVDENT | PIMTPNGAAG | VPADDPRVQE | RRVRSQRHRN |
| 250 | 260 | 270 | 280 | 290 | 300 |
| YMSRTHLHTP | PDLPEGYEQR | TTQQGQVYFL | HTQTGVSTWH | DPRVPRDLSN | VNCEELGPLP |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PGWEIRNTAT | GRVYFVDHNN | RTTQFTDPRL | SANLHLVLNP | SPNGSRAAVE | AQSSSRPGQL |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KEQAQSVVSP | GNLPEDPECL | TVPKYKRDLV | QKLKILRQEL | SQQQPQAGHC | RIEVSREEIF |
| 430 | 440 | 450 | 460 | 470 | 480 |
| EESYRQVMKM | RPKDLWKRLM | VKFRGEEGLD | YGGVAREWLY | LLSHEMLNPY | YGLFQYSRDD |
| 490 | 500 | 510 | 520 | 530 | 540 |
| IYTLQINPDS | AVNPEHLSYF | HFVGRIMGMA | VFHGHYIDGG | FTLPFYKQLL | GKPITLDDME |
| 550 | 560 | 570 | 580 | 590 | 600 |
| SVDPDLHNSL | VWILDNDITG | VLDHTFCVEH | NAYGEIIQHE | LKPNGKSIPV | TQDTKKEYVR |
| 610 | 620 | 630 | 640 | 650 | 660 |
| LYVNWRFLRG | IEAQFLALQK | GFNEVIPQHL | LKAFDEKELE | LIVCGLGKID | INDWKSNTRL |
| 670 | 680 | 690 | 700 | 710 | 720 |
| KHCTPDSNIV | KWFWRAVESY | DEERRARLLQ | FVTGSSRVPL | QGFKALQGAA | GPRLFTIHQI |
| 730 | 740 | 750 | 760 | ||
| DASTNNLPKA | HTCFNRIDIP | PYESYDKLYD | KLLTAIEETC | GFAVE |