A7ZPL3
Gene name |
zipA |
Protein name |
Cell division protein ZipA |
Names |
|
Species |
Escherichia coli O139:H28 (strain E24377A / ETEC) |
KEGG Pathway |
ecw:EcE24377A_2699 |
EC number |
|
Protein Class |
|
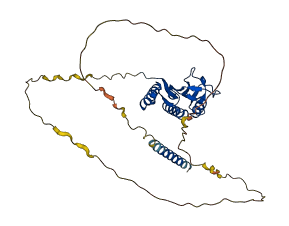
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for A7ZPL3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A7ZPL3-F1 | Predicted | AlphaFoldDB |
No variants for A7ZPL3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for A7ZPL3 | |||||
No associated diseases with A7ZPL3
8 regional properties for A7ZPL3
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Cyclic nucleotide-binding domain | 137 - 253 | IPR000595-1 |
| domain | Cyclic nucleotide-binding domain | 255 - 376 | IPR000595-2 |
| domain | cAMP-dependent protein kinase regulatory subunit, dimerization-anchoring domain | 25 - 62 | IPR003117 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 164 - 180 | IPR018488-1 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 200 - 217 | IPR018488-2 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 282 - 298 | IPR018488-3 |
| conserved_site | Cyclic nucleotide-binding, conserved site | 324 - 341 | IPR018488-4 |
| domain | RIbeta, dimerization/docking domain | 10 - 64 | IPR042818 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cell division site | The eventual plane of cell division (also known as cell cleavage or cytokinesis) in a dividing cell. In Eukaryotes, the cleavage apparatus, composed of septin structures and the actomyosin contractile ring, forms along this plane, and the mitotic, or meiotic, spindle is aligned perpendicular to the division plane. In bacteria, the cell division site is generally located at mid-cell and is the site at which the cytoskeletal structure, the Z-ring, assembles. |
| integral component of plasma membrane | The component of the plasma membrane consisting of the gene products and protein complexes having at least some part of their peptide sequence embedded in the hydrophobic region of the membrane. |
No GO annotations of molecular function
| Name | Definition |
|---|---|
| No GO annotations for molecular function |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| cell septum assembly | The assembly and arrangement of a cellular component that is composed of peptidoglycan and often chitin in addition to other materials and usually forms perpendicular to the long axis of a cell or hypha. It grows centripetally from the cell wall to the center of the cell and often functions in the compartmentalization of a cell into two daughter cells. |
| FtsZ-dependent cytokinesis | A cytokinesis process that involves a set of conserved proteins including FtsZ, and results in the formation of two similarly sized and shaped cells. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MMQDLRLILI | IVGAIAIIAF | LVHGFWTSRK | ERSSMFRDRP | LKRMKSKRDD | DSYDEDVEDD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| EGVGEVRVHR | VNHAPANAQE | HEAARPSPQH | QYQPPYASAQ | PRQPVQQPPE | AQVPPQHAPR |
| 130 | 140 | 150 | 160 | 170 | 180 |
| PAQPVQQPAY | QPQPEQPLQQ | PVSPQVAPAP | QPVHSAPQPA | QQAFQPAEPV | AAPQPEPVAE |
| 190 | 200 | 210 | 220 | 230 | 240 |
| PAPVMDKPKR | KEAVIIMNVA | AHHGSELNGE | LLLNSIQQAG | FIFGDMNIYH | RHLSPDGSGP |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ALFSLANMVK | PGTFDPEMKD | FTTPGVTIFM | QVPSYGDELQ | NFKLMLQSAQ | HIADEVGGVV |
| 310 | 320 | ||||
| LDDQRRMMTP | QKLREYQDII | REVKDANA |