A7E2Y1
Gene name |
MYH7B (KIAA1512) |
Protein name |
Myosin-7B |
Names |
Antigen MLAA-21 , Myosin cardiac muscle beta chain , Myosin heavy chain 7B, cardiac muscle beta isoform , Slow A MYH14 |
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:57644 |
EC number |
|
Protein Class |
|
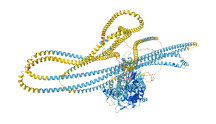
Descriptions
Autoinhibitory domains (AIDs)
Target domain |
120-828 (Myosin head, motor domain) |
Relief mechanism |
Partner binding |
Assay |
|
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for A7E2Y1
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A7E2Y1-F1 | Predicted | AlphaFoldDB |
8 variants for A7E2Y1
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs17092199 VAR_046359 |
70 | P>T | No |
UniProt dbSNP |
|
|
VAR_046360 rs754511 |
501 | F>Y | No |
UniProt dbSNP |
|
|
rs3746442 VAR_046361 |
780 | P>S | No |
UniProt dbSNP |
|
|
VAR_054814 rs2425015 |
1007 | E>K | No |
UniProt dbSNP |
|
|
VAR_046362 rs3746435 |
1552 | K>N | No |
UniProt dbSNP |
|
|
rs6060147 VAR_046363 |
1581 | A>V | No |
UniProt dbSNP |
|
|
VAR_046364 rs6060148 |
1698 | Q>R | No |
UniProt dbSNP |
|
|
rs7273482 VAR_046365 |
1917 | V>E | No |
UniProt dbSNP |
No associated diseases with A7E2Y1
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it and attached to it. |
| myosin filament | A supramolecular fiber containing myosin heavy chains, plus associated light chains and other proteins, in which the myosin heavy chains are arranged into a filament. |
| myosin II complex | A myosin complex containing two class II myosin heavy chains, two myosin essential light chains and two myosin regulatory light chains. Also known as classical myosin or conventional myosin, the myosin II class includes the major muscle myosin of vertebrate and invertebrate muscle, and is characterized by alpha-helical coiled coil tails that self assemble to form a variety of filament structures. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| actin filament binding | Binding to an actin filament, also known as F-actin, a helical filamentous polymer of globular G-actin subunits. |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| microfilament motor activity | A motor activity that generates movement along a microfilament, driven by ATP hydrolysis. |
No GO annotations of biological process
| Name | Definition |
|---|---|
| No GO annotations for biological process |
46 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9BE40 | MYH1 | Myosin-1 | Bos taurus (Bovine) | SS |
| Q9BE41 | MYH2 | Myosin-2 | Bos taurus (Bovine) | SS |
| Q27991 | MYH10 | Myosin-10 | Bos taurus (Bovine) | SS |
| Q9BE39 | MYH7 | Myosin-7 | Bos taurus (Bovine) | SS |
| P10587 | MYH11 | Myosin-11 | Gallus gallus (Chicken) | SS |
| P14105 | MYH9 | Myosin-9 | Gallus gallus (Chicken) | SS |
| P02565 | MYH1B | Myosin-1B | Gallus gallus (Chicken) | SS |
| P13538 | Myosin heavy chain, skeletal muscle, adult | Gallus gallus (Chicken) | SS | |
| Q99323 | zip | Myosin heavy chain, non-muscle | Drosophila melanogaster (Fruit fly) | SS |
| P05661 | Mhc | Myosin heavy chain, muscle | Drosophila melanogaster (Fruit fly) | SS |
| P11055 | MYH3 | Myosin-3 | Homo sapiens (Human) | SS |
| P12882 | MYH1 | Myosin-1 | Homo sapiens (Human) | SS |
| P12883 | MYH7 | Myosin-7 | Homo sapiens (Human) | EV |
| P13533 | MYH6 | Myosin-6 | Homo sapiens (Human) | SS |
| P13535 | MYH8 | Myosin-8 | Homo sapiens (Human) | SS |
| Q9UKX3 | MYH13 | Myosin-13 | Homo sapiens (Human) | SS |
| Q9Y2K3 | MYH15 | Myosin-15 | Homo sapiens (Human) | SS |
| Q9Y623 | MYH4 | Myosin-4 | Homo sapiens (Human) | SS |
| Q9UKX2 | MYH2 | Myosin-2 | Homo sapiens (Human) | SS |
| P35580 | MYH10 | Myosin-10 | Homo sapiens (Human) | SS |
| P35749 | MYH11 | Myosin-11 | Homo sapiens (Human) | SS |
| P35579 | MYH9 | Myosin-9 | Homo sapiens (Human) | SS |
| Q7Z406 | MYH14 | Myosin-14 | Homo sapiens (Human) | SS |
| Q8VDD5 | Myh9 | Myosin-9 | Mus musculus (Mouse) | SS |
| Q5SX39 | Myh4 | Myosin-4 | Mus musculus (Mouse) | SS |
| P13542 | Myh8 | Myosin-8 | Mus musculus (Mouse) | SS |
| Q02566 | Myh6 | Myosin-6 | Mus musculus (Mouse) | SS |
| O08638 | Myh11 | Myosin-11 | Mus musculus (Mouse) | SS |
| Q61879 | Myh10 | Myosin-10 | Mus musculus (Mouse) | SS |
| Q91Z83 | Myh7 | Myosin-7 | Mus musculus (Mouse) | SS |
| Q6URW6 | Myh14 | Myosin-14 | Mus musculus (Mouse) | SS |
| P13541 | Myh3 | Myosin-3 | Mus musculus (Mouse) | SS |
| Q5SX40 | Myh1 | Myosin-1 | Mus musculus (Mouse) | SS |
| A2AQP0 | Myh7b | Myosin-7B | Mus musculus (Mouse) | SS |
| P79293 | MYH7 | Myosin-7 | Sus scrofa (Pig) | SS |
| Q9TV63 | MYH2 | Myosin-2 | Sus scrofa (Pig) | SS |
| P12847 | Myh3 | Myosin-3 | Rattus norvegicus (Rat) | SS |
| P02563 | Myh6 | Myosin-6 | Rattus norvegicus (Rat) | SS |
| P02564 | Myh7 | Myosin-7 | Rattus norvegicus (Rat) | SS |
| Q62812 | Myh9 | Myosin-9 | Rattus norvegicus (Rat) | SS |
| Q29RW1 | Myh4 | Myosin-4 | Rattus norvegicus (Rat) | SS |
| Q9JLT0 | Myh10 | Myosin-10 | Rattus norvegicus (Rat) | SS |
| P02566 | unc-54 | Myosin-4 | Caenorhabditis elegans | SS |
| P02567 | myo-1 | Myosin-1 | Caenorhabditis elegans | SS |
| P12844 | myo-3 | Myosin-3 | Caenorhabditis elegans | SS |
| P12845 | myo-2 | Myosin-2 | Caenorhabditis elegans | SS |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSGNKRGSRA | SCPHRGAECL | LPWAALNLQG | FQLLLLHPSA | TAMMDVSELG | ESARYLRQGY |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QEMTKVHTIP | WDGKKRVWVP | DEQDAYVEAE | VKSEATGGRV | TVETKDQKVL | MVREAELQPM |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NPPRFDLLED | MAMMTHLNEA | SVLHNLRQRY | ARWMIYTYSG | LFCVTINPYK | WLPVYTASVV |
| 190 | 200 | 210 | 220 | 230 | 240 |
| AAYKGKRRSD | SPPHIYAVAD | NAYNDMLRNR | DNQSMLITGE | SGAGKTVNTK | RVIQYFAIVA |
| 250 | 260 | 270 | 280 | 290 | 300 |
| ALGDGPGKKA | QFLATKTGGT | LEDQIIEANP | AMEAFGNAKT | LRNDNSSRFG | KFIRIHFGPS |
| 310 | 320 | 330 | 340 | 350 | 360 |
| GKLASADIDS | YLLEKSRVIF | QLPGERSYHV | YYQILSGRKP | ELQDMLLLSM | NPYDYHFCSQ |
| 370 | 380 | 390 | 400 | 410 | 420 |
| GVITVDNMND | GEELIATDHA | MDILGFSVDE | KCACYKIVGA | LLHFGNMKFK | QKQREEQAEA |
| 430 | 440 | 450 | 460 | 470 | 480 |
| DGTESADKAA | YLMGVSSGDL | LKGLLHPRVR | VGNEYVTKGQ | SVEQVVFAVG | ALAKATYDRL |
| 490 | 500 | 510 | 520 | 530 | 540 |
| FRWLVSRINQ | TLDTKLPRQF | FIGVLDIAGF | EIFEFNSFEQ | LCINFTNEKL | QQFFNQHMFV |
| 550 | 560 | 570 | 580 | 590 | 600 |
| LEQEEYKREG | IDWVFIDFGL | DLQPCIDLIE | KPLGILSILE | EECMFPKASD | ASFRAKLYDN |
| 610 | 620 | 630 | 640 | 650 | 660 |
| HAGKSPNFQQ | PRPDKKRKYQ | AHFEVVHYAG | VVPYSIVGWL | EKNKDPLNET | VVPIFQKSQN |
| 670 | 680 | 690 | 700 | 710 | 720 |
| RLLATLYENY | AGSCSTEPPK | SGVKEKRKKA | ASFQTVSQLH | KENLNKLMTN | LRATQPHFVR |
| 730 | 740 | 750 | 760 | 770 | 780 |
| CIVPNENKTP | GVMDAFLVLH | QLRCNGVLEG | IRICRQGFPN | RLLYTDFRQR | YRILNPSAIP |
| 790 | 800 | 810 | 820 | 830 | 840 |
| DDTFMDSRKA | TEKLLGSLDL | DHTQYQFGHT | KVFFKAGLLG | VLEELRDQRL | AKVLTLLQAR |
| 850 | 860 | 870 | 880 | 890 | 900 |
| SRGRLMRLEY | QRLLGGRDAL | FTIQWNIRAF | NAVKNWSWMK | LFFKMKPLLR | SAQAEEELAA |
| 910 | 920 | 930 | 940 | 950 | 960 |
| LRAELRGLRG | ALAAAEAKRQ | ELEETHVSIT | QEKNDLALQL | QAEQDNLADA | EERCHLLIKS |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| KVQLEGKVKE | LSERLEDEEE | VNADLAARRR | KLEDECTELK | KDIDDLELTL | AKAEKEKQAT |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| ENKVKNLTEE | MAALDESVAR | LTKEKKALQE | AHQQALGDLQ | AEEDRVSALT | KAKLRLEQQV |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| EDLECSLEQE | KKLRMDTERA | KRKLEGDLKL | TQESVADAAQ | DKQQLEEKLK | KKDSELSQLS |
| 1150 | 1160 | 1170 | 1180 | 1190 | 1200 |
| LRVEDEQLLG | AQMQKKIKEL | QARAEELEEE | LEAERAARAR | VEKQRAEAAR | ELEELSERLE |
| 1210 | 1220 | 1230 | 1240 | 1250 | 1260 |
| EAGGASAGQR | EGCRKREAEL | GRLRRELEEA | ALRHEATVAA | LRRKQAEGAA | ELGEQVDSLQ |
| 1270 | 1280 | 1290 | 1300 | 1310 | 1320 |
| RVRQKLEKEK | SELRMEVDDL | AANVETLTRA | KASAEKLCRT | YEDQLSEAKI | KVEELQRQLA |
| 1330 | 1340 | 1350 | 1360 | 1370 | 1380 |
| DASTQRGRLQ | TESGELSRLL | EEKECLISQL | SRGKALAAQS | LEELRRQLEE | ESKAKSALAH |
| 1390 | 1400 | 1410 | 1420 | 1430 | 1440 |
| AVQALRHDCD | LLREQHEEEA | EAQAELQRLL | SKANAEVAQW | RSKYEADAIQ | RTEELEEAKK |
| 1450 | 1460 | 1470 | 1480 | 1490 | 1500 |
| KLALRLQEAE | EGVEAANAKC | SSLEKAKLRL | QTESEDVTLE | LERATSAAAA | LDKKQRHLER |
| 1510 | 1520 | 1530 | 1540 | 1550 | 1560 |
| ALEERRRQEE | EMQRELEAAQ | RESRGLGTEL | FRLRHGHEEA | LEALETLKRE | NKNLQEEISD |
| 1570 | 1580 | 1590 | 1600 | 1610 | 1620 |
| LTDQVSLSGK | SIQELEKTKK | ALEGEKSEIQ | AALEEAEGAL | ELEETKTLRI | QLELSQVKAE |
| 1630 | 1640 | 1650 | 1660 | 1670 | 1680 |
| VDRKLAEKDE | ECANLRRNHQ | RAVESLQASL | DAETRARNEA | LRLKKKMEGD | LNDLELQLGH |
| 1690 | 1700 | 1710 | 1720 | 1730 | 1740 |
| ATRQATEAQA | ATRLMQAQLK | EEQAGRDEEQ | RLAAELHEQA | QALERRASLL | AAELEELRAA |
| 1750 | 1760 | 1770 | 1780 | 1790 | 1800 |
| LEQGERSRRL | AEQELLEATE | RLNLLHSQNT | GLLNQKKKLE | ADLAQLSGEV | EEAAQERREA |
| 1810 | 1820 | 1830 | 1840 | 1850 | 1860 |
| EEKAKKAITD | AAMMAEELKK | EQDTSAHLER | MKKTLEQTVR | ELQARLEEAE | QAALRGGKKQ |
| 1870 | 1880 | 1890 | 1900 | 1910 | 1920 |
| VQKLEAKVRE | LEAELDAEQK | KHAEALKGVR | KHERRVKELA | YQAEEDRKNL | ARMQDLVDKL |
| 1930 | 1940 | 1950 | 1960 | 1970 | 1980 |
| QSKVKSYKRQ | FEEAEQQANT | NLAKYRKAQH | ELDDAEERAD | MAETQANKLR | ARTRDALGPK |
| HKE |