A4IID4
Gene name |
pi4kb (pik4cb) |
Protein name |
Phosphatidylinositol 4-kinase beta |
Names |
PI4K-beta, PI4Kbeta, PtdIns 4-kinase beta |
Species |
Xenopus tropicalis (Western clawed frog) (Silurana tropicalis) |
KEGG Pathway |
xtr:100124755 |
EC number |
2.7.1.67: Phosphotransferases with an alcohol group as acceptor |
Protein Class |
|
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
679-701 (Activation loop from InterPro)
Target domain |
519-806 (Catalytic domain of Type III Phosphoinositide 4-kinase beta) |
Relief mechanism |
|
Assay |
|
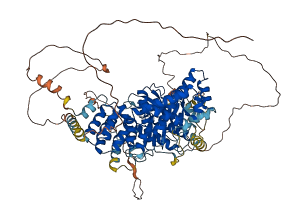
Autoinhibited structure

Activated structure

1 structures for A4IID4
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A4IID4-F1 | Predicted | AlphaFoldDB |
No variants for A4IID4
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| No variants for A4IID4 | |||||
No associated diseases with A4IID4
4 regional properties for A4IID4
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Phosphatidylinositol 3-/4-kinase, catalytic domain | 525 - 804 | IPR000403 |
| domain | Phosphoinositide 3-kinase, accessory (PIK) domain | 55 - 247 | IPR001263 |
| conserved_site | Phosphatidylinositol 3/4-kinase, conserved site | 553 - 567 | IPR018936-1 |
| conserved_site | Phosphatidylinositol 3/4-kinase, conserved site | 646 - 666 | IPR018936-2 |
Functions
| Description | ||
|---|---|---|
| EC Number | 2.7.1.67 | Phosphotransferases with an alcohol group as acceptor |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
4 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| membrane | A lipid bilayer along with all the proteins and protein complexes embedded in it an attached to it. |
| mitochondrial outer membrane | The outer, i.e. cytoplasm-facing, lipid bilayer of the mitochondrial envelope. |
| rough endoplasmic reticulum membrane | The lipid bilayer surrounding the rough endoplasmic reticulum. |
3 GO annotations of molecular function
| Name | Definition |
|---|---|
| 1-phosphatidylinositol 4-kinase activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol + ATP = 1-phosphatidyl-1D-myo-inositol 4-phosphate + ADP + 2 H(+). |
| ATP binding | Binding to ATP, adenosine 5'-triphosphate, a universally important coenzyme and enzyme regulator. |
| phosphatidylinositol kinase activity | Catalysis of the reaction: ATP + a phosphatidylinositol = ADP + a phosphatidylinositol phosphate. |
3 GO annotations of biological process
| Name | Definition |
|---|---|
| phosphatidylinositol phosphate biosynthetic process | The chemical reactions and pathways resulting in the formation of phosphatidylinositol phosphate. |
| phosphatidylinositol-mediated signaling | The series of molecular signals in which a cell uses a phosphatidylinositol-mediated signaling to convert a signal into a response. Phosphatidylinositols include phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| phosphorylation | The process of introducing a phosphate group into a molecule, usually with the formation of a phosphoric ester, a phosphoric anhydride or a phosphoric amide. |
6 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P39104 | PIK1 | Phosphatidylinositol 4-kinase PIK1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| O02810 | PI4KB | Phosphatidylinositol 4-kinase beta | Bos taurus (Bovine) | PR |
| Q9UBF8 | PI4KB | Phosphatidylinositol 4-kinase beta | Homo sapiens (Human) | PR |
| Q8BKC8 | Pi4kb | Phosphatidylinositol 4-kinase beta | Mus musculus (Mouse) | PR |
| O08561 | Pi4kb | Phosphatidylinositol 4-kinase beta | Rattus norvegicus (Rat) | PR |
| Q49GP3 | pi4kb | Phosphatidylinositol 4-kinase beta | Danio rerio (Zebrafish) (Brachydanio rerio) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MGDTMVEPVP | AKLSDPTLVL | RGNGGSPLCV | ITEGVGEAQM | VIDPDVAEKA | CQDVLDKVKL |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IRGSSAESLD | KIDGSDTGDG | GSLANGDAGP | RHSESCGPPV | SASRITEEEE | SLIDINSVKS |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ARRRQKNNSA | KQSWLLRLFE | CKLFDVSMAI | SYLYNSKEPG | VQAYIGNRLF | CFRYEDVDFY |
| 190 | 200 | 210 | 220 | 230 | 240 |
| LPQLLNMYIH | MDEDVGDAIK | PYVVHRCRQS | INFSLQCAWL | LGAYSSDMHI | STQRHSRGTK |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LRKLILSDEL | KPAHKKREIP | PLSLAPDTGL | SPSKRTHQRS | KSDATVSISL | SSNLKRTSSN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PKVENDDEPV | RLAPEREFIK | SLMAIGKRLA | TLPTKEQKTQ | RLISELSLLN | HKLPARVWLP |
| 370 | 380 | 390 | 400 | 410 | 420 |
| TAGFDHHVVR | VPHTQAVVLN | SKDKAPYLIY | VEVLECENFE | TSLVPVRIPE | NRIRSTRSVE |
| 430 | 440 | 450 | 460 | 470 | 480 |
| NLPECGITHE | QRASSFTTVP | NYDNDDEAWS | VDDIGELQVE | LPELHTNSCD | NISQFSVDSI |
| 490 | 500 | 510 | 520 | 530 | 540 |
| TSQESKDPVF | IAAGDIRRRL | SEQLAHTPTT | FRRDPEDPSA | VALKEPWQEK | VRRIREGSPY |
| 550 | 560 | 570 | 580 | 590 | 600 |
| GHFPNWRLLS | VIVKCGDDLR | QELLAYQVLK | QLQSIWESER | VPLWIRPYKI | LVISADSGMI |
| 610 | 620 | 630 | 640 | 650 | 660 |
| EPVVNAVSIH | QVKKQSQLSL | LDYFLQEHGS | CTTEAFLTAQ | RNFVQSCAGY | CLVCYLLQVK |
| 670 | 680 | 690 | 700 | 710 | 720 |
| DRHNGNILLD | AEGHIIHIDF | GFILSSSPRN | LGFETSAFKL | TSEFVDVMGG | LNGDMFNYYK |
| 730 | 740 | 750 | 760 | 770 | 780 |
| MLMLQGLIAA | RKHMDRVVQI | VEIMQQGSQL | PCFHGSSTIR | NLKERFHMNM | TEEQLQVLVE |
| 790 | 800 | ||||
| QMVDGSMRSI | TTKLYDGFQY | LTNGIM |