A4D1S5
Gene name |
RAB19 (RAB19B) |
Protein name |
Ras-related protein Rab-19 |
Names |
|
Species |
Homo sapiens (Human) |
KEGG Pathway |
hsa:401409 |
EC number |
|
Protein Class |
|
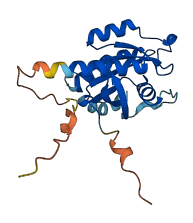
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for A4D1S5
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A4D1S5-F1 | Predicted | AlphaFoldDB |
210 variants for A4D1S5
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
|
rs776077715 CA4514154 |
2 | H>N | No |
ClinGen ExAC gnomAD |
|
|
rs776077715 CA369516005 |
2 | H>Y | No |
ClinGen ExAC gnomAD |
|
|
rs1311611197 CA369516063 |
6 | S>L | No |
ClinGen TOPMed |
|
|
rs1230512555 CA369516077 |
7 | A>G | No |
ClinGen TOPMed |
|
|
CA4514157 rs774958091 |
10 | A>G | No |
ClinGen ExAC gnomAD |
|
|
rs769494285 CA4514156 |
10 | A>T | No |
ClinGen ExAC gnomAD |
|
|
rs1365712877 CA369516128 |
11 | D>V | No |
ClinGen TOPMed |
|
|
CA369516194 rs1434754327 |
15 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
rs1296568020 CA369516184 |
15 | D>Y | No |
ClinGen TOPMed |
|
|
CA369516233 rs1451888562 |
18 | F>S | No |
ClinGen gnomAD |
|
|
rs764459628 CA4514159 |
19 | K>E | No |
ClinGen ExAC gnomAD |
|
|
CA4514161 rs762018113 |
22 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs767923063 CA4514162 |
23 | I>F | No |
ClinGen ExAC gnomAD |
|
|
rs140181115 CA4514163 |
23 | I>T | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs767923063 CA369516296 |
23 | I>V | No |
ClinGen ExAC gnomAD |
|
|
rs974688905 CA167996331 |
24 | G>A | No |
ClinGen TOPMed |
|
|
CA369516316 rs1432140332 |
25 | D>E | No |
ClinGen TOPMed |
|
|
rs367821693 CA4514164 |
27 | N>S | Variant assessed as Somatic; 4.619e-05 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA4514166 rs372047383 |
31 | T>M | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs1182607982 CA369516397 |
32 | C>Y | No |
ClinGen TOPMed |
|
|
CA369516417 rs1184215603 |
33 | V>L | No |
ClinGen gnomAD |
|
|
CA369516430 rs770838607 |
34 | V>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514170 rs770838607 |
34 | V>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA167996368 rs962861019 |
35 | Q>* | No |
ClinGen Ensembl |
|
|
CA167996380 rs972934385 |
35 | Q>P | No |
ClinGen Ensembl |
|
|
CA4514172 rs745711953 |
39 | S>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA167996394 rs955778115 |
41 | V>F | No |
ClinGen TOPMed |
|
|
rs1368428864 CA369516635 |
42 | Y>H | No |
ClinGen gnomAD |
|
|
CA167996411 rs939506920 |
43 | T>N | No |
ClinGen TOPMed |
|
|
rs1403756604 CA369516665 COSM1729807 |
43 | T>S | liver [Cosmic] | No |
ClinGen cosmic curated gnomAD |
|
COSM3411662 CA369516816 rs1164371377 |
49 | T>M | central_nervous_system Variant assessed as Somatic; 4.619e-05 impact. [Cosmic, NCI-TCGA] | No |
ClinGen cosmic curated NCI-TCGA gnomAD |
|
rs148552166 CA4514174 |
50 | I>T | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4514176 rs768219369 |
52 | V>E | No |
ClinGen ExAC |
|
|
CA4514175 rs748738710 |
52 | V>L | No |
ClinGen ExAC gnomAD |
|
|
CA4514177 rs773801299 |
53 | D>N | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA369516963 rs1585412981 |
55 | T>I | No |
ClinGen Ensembl |
|
|
rs369833403 CA4514179 |
56 | V>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA4514180 rs773684234 |
57 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs373562675 CA4514181 |
57 | R>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs373562675 CA369517006 |
57 | R>L | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA369517067 rs866283902 |
60 | D>E | No |
ClinGen gnomAD |
|
|
CA369517055 rs1307835780 |
60 | D>H | No |
ClinGen gnomAD |
|
|
rs546014360 CA4514184 |
61 | I>M | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs754122207 CA4514183 |
61 | I>S | No |
ClinGen ExAC gnomAD |
|
|
CA369517098 rs754122207 |
61 | I>T | No |
ClinGen ExAC gnomAD |
|
|
CA834209556 rs1165798030 |
63 | G>* | No |
ClinGen TOPMed |
|
|
CA167996479 rs935164126 |
63 | G>D | No |
ClinGen TOPMed gnomAD |
|
|
rs752775630 CA4514186 |
63 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs935164126 CA369517134 |
63 | G>V | No |
ClinGen TOPMed gnomAD |
|
|
COSM1086223 CA4514188 rs564143161 |
64 | K>T | endometrium [Cosmic] | No |
ClinGen cosmic curated 1000Genomes ExAC TOPMed gnomAD |
|
CA369517180 rs1451718336 |
66 | V>L | No |
ClinGen TOPMed gnomAD |
|
|
CA369517183 rs1451718336 |
66 | V>M | No |
ClinGen TOPMed gnomAD |
|
|
rs1243681549 CA369517210 |
67 | K>R | No |
ClinGen TOPMed |
|
|
CA168000254 rs1011805884 |
68 | M>T | No |
ClinGen Ensembl |
|
|
rs201141254 CA168000262 |
69 | Q>* | No |
ClinGen gnomAD |
|
|
rs756849367 CA4514264 |
71 | W>* | No |
ClinGen ExAC |
|
|
rs990303151 CA168000269 |
72 | D>Y | No |
ClinGen TOPMed |
|
|
rs1307785364 CA369519674 |
73 | T>I | No |
ClinGen Ensembl |
|
|
CA168000273 rs1043675447 |
74 | A>T | No |
ClinGen gnomAD |
|
|
CA369519730 rs1397907950 |
76 | Q>K | No |
ClinGen gnomAD |
|
|
rs367625241 CA168000276 |
76 | Q>R | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen ESP NCI-TCGA TOPMed |
|
rs1449804802 CA369519771 |
77 | E>* | No |
ClinGen TOPMed |
|
|
CA369519809 rs1340028480 |
78 | R>C | No |
ClinGen TOPMed gnomAD |
|
|
rs201700792 CA4514266 |
78 | R>H | No |
ClinGen ExAC gnomAD |
|
|
rs142256906 CA4514267 |
79 | F>S | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
rs576701732 CA4514268 |
80 | R>C | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA4514269 rs146219058 |
80 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA369519881 rs146219058 |
80 | R>P | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4514271 rs138951503 |
82 | I>V | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4514272 rs748172408 |
83 | T>N | No |
ClinGen ExAC gnomAD |
|
|
CA168000313 rs200166138 |
84 | Q>* | No |
ClinGen ESP TOPMed gnomAD |
|
|
CA369519998 rs1253466393 |
84 | Q>R | No |
ClinGen TOPMed gnomAD |
|
|
rs1441421590 CA369520065 |
85 | S>R | No |
ClinGen gnomAD |
|
|
CA4514273 rs148867635 |
86 | Y>* | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs1337185398 CA369520092 |
86 | Y>F | No |
ClinGen TOPMed gnomAD |
|
|
CA369520073 rs1291109526 |
86 | Y>H | No |
ClinGen TOPMed gnomAD |
|
|
rs771078209 CA4514274 |
86 | Y>KFYINSMRI* | No |
ClinGen ExAC |
|
|
CA369520082 rs1337185398 |
86 | Y>S | No |
ClinGen TOPMed gnomAD |
|
|
rs1585419271 CA369520149 |
87 | Y>S | No |
ClinGen Ensembl |
|
|
CA369520219 rs1239263920 |
88 | R>C | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA369520218 rs1239263920 |
88 | R>G | No |
ClinGen TOPMed gnomAD |
|
|
CA369520220 rs1447472611 |
88 | R>H | No |
ClinGen gnomAD |
|
|
CA369520322 rs375880877 |
91 | H>Q | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA369520339 rs189143966 |
92 | A>S | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514276 rs189143966 |
92 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA4514278 rs367914337 |
94 | I>F | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4514277 rs367914337 |
94 | I>V | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA369520462 rs1240429143 |
95 | I>N | No |
ClinGen TOPMed |
|
|
rs377517143 CA168000337 |
96 | A>D | No |
ClinGen Ensembl |
|
|
CA4514281 rs751270524 |
96 | A>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514282 rs757001951 |
97 | Y>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA369520619 rs1434904933 |
98 | D>E | No |
ClinGen TOPMed gnomAD |
|
|
CA369520663 rs1485406638 |
100 | T>I | No |
ClinGen TOPMed gnomAD |
|
|
CA369520655 rs1485406638 |
100 | T>N | No |
ClinGen TOPMed gnomAD |
|
|
rs1485406638 CA369520661 |
100 | T>S | No |
ClinGen TOPMed gnomAD |
|
|
CA4514285 rs1554441478 |
101 | R>Q | No |
ClinGen Ensembl |
|
|
CA4514284 rs527792086 |
101 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514288 rs570778385 |
102 | R>L | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs570778385 CA4514289 |
102 | R>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514287 rs552497522 |
102 | R>W | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514290 rs755346342 |
103 | S>F | No |
ClinGen ExAC gnomAD |
|
|
CA168000399 rs962071591 |
104 | T>M | No |
ClinGen gnomAD |
|
|
CA168000404 rs369704079 |
106 | E>K | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA TOPMed gnomAD |
|
CA4514293 rs772398775 |
110 | H>N | No |
ClinGen ExAC gnomAD |
|
|
rs747154819 CA369520939 TCGA novel |
110 | H>Q | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
NCI-TCGA ClinGen ExAC TOPMed gnomAD |
|
rs550527590 CA4514294 |
110 | H>R | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA4514296 rs201226450 |
111 | W>R | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514299 rs762778683 |
115 | I>L | No |
ClinGen ExAC gnomAD |
|
|
CA369521061 rs1294424120 |
115 | I>T | No |
ClinGen TOPMed gnomAD |
|
|
CA369521056 rs762778683 |
115 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA4514300 rs763986254 |
116 | E>K | No |
ClinGen ExAC |
|
|
CA4514301 rs182029506 |
118 | Y>* | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA369521195 rs1385555256 |
119 | G>A | No |
ClinGen gnomAD |
|
|
CA369521192 rs1385555256 |
119 | G>E | No |
ClinGen gnomAD |
|
|
CA168000444 rs986158378 |
120 | A>P | No |
ClinGen TOPMed |
|
|
rs767237902 CA4514303 |
121 | A>P | No |
ClinGen ExAC gnomAD |
|
|
CA168000460 rs910736617 |
121 | A>V | No |
ClinGen TOPMed gnomAD |
|
|
rs746761201 CA168000464 |
122 | N>S | No |
ClinGen TOPMed |
|
|
CA168000465 rs375148253 |
123 | V>M | No |
ClinGen ESP TOPMed |
|
|
rs1428377700 CA369521287 |
124 | V>F | No |
ClinGen TOPMed |
|
|
rs1189197549 CA369521294 |
125 | I>L | No |
ClinGen TOPMed |
|
| TCGA novel | 125 | I>V | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA369521343 rs1269620334 |
127 | L>M | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
rs183591043 CA4514304 |
127 | L>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs183591043 CA4514305 |
127 | L>Q | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA369521346 rs1269620334 |
127 | L>V | No |
ClinGen gnomAD |
|
|
CA369521383 rs1214664750 |
128 | I>T | No |
ClinGen gnomAD |
|
|
rs1251580589 CA369523694 |
130 | N>K | No |
ClinGen TOPMed |
|
|
rs774254709 CA4514319 |
132 | C>R | No |
ClinGen ExAC gnomAD |
|
|
CA4514320 rs761696770 |
132 | C>Y | No |
ClinGen ExAC gnomAD |
|
|
CA168009280 rs955985079 |
133 | D>A | No |
ClinGen TOPMed |
|
|
rs771572401 CA4514321 |
134 | L>P | No |
ClinGen ExAC gnomAD |
|
|
rs760033770 CA4514323 |
135 | W>* | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514322 rs772928313 |
135 | W>R | No |
ClinGen ExAC |
|
|
rs375344241 CA4514324 |
136 | E>G | No |
ClinGen ESP ExAC gnomAD |
|
|
rs1274483320 CA369523787 |
137 | K>E | No |
ClinGen TOPMed |
|
|
CA4514326 rs760017924 |
138 | R>Q | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514325 rs753408586 |
138 | R>W | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs1391195670 CA369523832 |
140 | V>A | No |
ClinGen gnomAD |
|
|
CA4514328 rs752965955 |
140 | V>I | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs898261798 CA168009312 |
141 | L>P | No |
ClinGen TOPMed |
|
|
CA4514330 rs777801769 |
142 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514333 rs570628599 |
143 | E>K | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
rs570628599 CA4514332 |
143 | E>Q | Variant assessed as Somatic; 4.621e-05 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA gnomAD |
|
CA4514334 rs745983601 |
145 | A>V | No |
ClinGen ExAC gnomAD |
|
|
CA4514335 rs768672408 |
147 | T>S | No |
ClinGen ExAC gnomAD |
|
|
rs747837185 CA4514337 |
152 | Y>C | No |
ClinGen ExAC gnomAD |
|
|
CA4514336 rs2948386 |
152 | Y>H | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs771901851 CA4514338 |
153 | G>S | No |
ClinGen ExAC TOPMed gnomAD |
|
|
rs772942378 CA4514339 |
155 | L>P | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514340 rs370140812 |
156 | A>D | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA4514343 rs568276754 |
157 | V>F | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514342 rs568276754 |
157 | V>I | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
rs765685964 CA4514344 |
158 | L>V | No |
ClinGen ExAC gnomAD |
|
|
CA369524936 rs1167249451 |
161 | S>C | No |
ClinGen gnomAD |
|
|
CA168009401 rs1040990196 |
161 | S>T | No |
ClinGen TOPMed gnomAD |
|
|
CA369524951 rs1419515548 |
162 | A>V | No |
ClinGen gnomAD |
|
|
rs763161215 CA4514346 |
163 | K>R | No |
ClinGen ExAC gnomAD |
|
|
CA4514347 rs764514529 |
165 | S>* | No |
ClinGen ExAC gnomAD |
|
|
rs751917314 CA4514348 |
167 | N>T | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514352 rs574686417 |
168 | I>T | No |
ClinGen ExAC TOPMed |
|
|
rs767881775 CA4514350 |
168 | I>V | No |
ClinGen ExAC gnomAD |
|
|
CA369525060 rs1437082644 |
169 | E>K | No |
ClinGen gnomAD |
|
|
rs371818531 CA4514356 |
172 | F>L | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514357 rs146383208 |
173 | V>M | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA369525187 rs1203247207 |
174 | L>P | No |
ClinGen gnomAD |
|
|
rs1269074412 CA369525202 |
175 | M>K | No |
ClinGen TOPMed gnomAD |
|
|
rs1279712789 CA369525198 |
175 | M>L | No |
ClinGen TOPMed |
|
|
rs1279712789 CA369525195 |
175 | M>V | No |
ClinGen TOPMed |
|
|
CA168009473 rs957491901 |
178 | E>* | No |
ClinGen Ensembl |
|
|
CA369525267 rs988823882 |
178 | E>G | No |
ClinGen gnomAD |
|
|
rs988823882 CA168009475 |
178 | E>V | No |
ClinGen gnomAD |
|
|
CA369525301 rs776290345 |
180 | I>M | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514361 rs745326291 |
181 | A>T | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ExAC NCI-TCGA TOPMed gnomAD |
|
rs554346458 CA4514362 |
181 | A>V | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
CA4514365 rs764549397 |
182 | R>C | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA4514366 rs558153548 |
182 | R>H | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen 1000Genomes ExAC NCI-TCGA TOPMed gnomAD |
|
CA369525327 rs558153548 |
182 | R>P | No |
ClinGen 1000Genomes ExAC TOPMed gnomAD |
|
|
rs762290218 CA4514367 |
184 | S>I | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 184 | S>N | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA4514368 rs1554443009 |
186 | H>P | No |
ClinGen Ensembl |
|
|
rs1458491429 CA369525456 |
188 | Y>C | No |
ClinGen gnomAD |
|
|
CA4514370 rs144022880 |
188 | Y>H | No |
ClinGen 1000Genomes ExAC gnomAD |
|
|
CA369525579 rs1315888324 |
191 | S>I | No |
ClinGen gnomAD |
|
|
rs1325660696 CA369525600 CA369525604 |
191 | S>R | No |
ClinGen gnomAD |
|
|
rs1411579062 CA369525636 |
192 | A>V | No |
ClinGen TOPMed |
|
|
CA168009531 rs992707189 |
193 | L>Q | No |
ClinGen TOPMed |
|
|
rs4585665 CA369525698 CA4514373 |
194 | N>K | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4514374 rs144362151 |
195 | G>S | Variant assessed as Somatic; 0.0 impact. [NCI-TCGA] | No |
ClinGen ESP ExAC NCI-TCGA TOPMed gnomAD |
|
CA168009596 rs777605625 |
198 | L>V | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA369525788 rs1223852264 |
199 | D>A | No |
ClinGen TOPMed |
|
| TCGA novel | 199 | D>E | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA369525782 rs1207086674 |
199 | D>H | No |
ClinGen gnomAD |
|
|
rs1272939081 CA369525820 |
201 | S>C | No |
ClinGen TOPMed |
|
|
rs757058982 CA4514378 |
202 | P>H | No |
ClinGen ExAC gnomAD |
|
| TCGA novel | 203 | V>A | Variant assessed as Somatic; impact. [NCI-TCGA] | No | NCI-TCGA |
|
CA369525856 rs144754394 |
203 | V>F | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
CA4514379 rs144754394 |
203 | V>I | No |
ClinGen 1000Genomes ESP ExAC TOPMed gnomAD |
|
|
rs147871231 CA369525901 CA4514380 |
205 | M>I | No |
ClinGen ESP ExAC TOPMed gnomAD |
|
|
CA369525896 rs1563076461 |
205 | M>T | No |
ClinGen Ensembl |
|
|
CA369525914 rs1197980303 |
206 | A>D | No |
ClinGen gnomAD |
|
|
rs1473339449 CA369525933 |
207 | Q>R | No |
ClinGen gnomAD |
|
|
CA369525946 rs1327142718 |
208 | G>S | Variant assessed as Somatic; impact. [NCI-TCGA] | No |
ClinGen NCI-TCGA gnomAD |
|
CA4514382 rs775034178 |
209 | P>L | No |
ClinGen ExAC gnomAD |
|
|
CA369525979 rs775034178 |
209 | P>R | No |
ClinGen ExAC gnomAD |
|
|
rs769155753 CA4514384 |
210 | S>G | No |
ClinGen ExAC TOPMed gnomAD |
|
|
CA369526002 rs1325441561 |
210 | S>I | No |
ClinGen TOPMed gnomAD |
|
|
rs774732290 CA4514385 |
210 | S>R | No |
ClinGen ExAC |
|
|
CA369526076 rs1363571185 |
214 | H>Y | No |
ClinGen TOPMed gnomAD |
|
|
rs1172427827 CA369526126 |
216 | T>S | No |
ClinGen TOPMed |
No associated diseases with A4D1S5
1 regional properties for A4D1S5
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Small GTP-binding protein domain | 16 - 175 | IPR005225 |
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| extracellular exosome | A vesicle that is released into the extracellular region by fusion of the limiting endosomal membrane of a multivesicular body with the plasma membrane. Extracellular exosomes, also simply called exosomes, have a diameter of about 40-100 nm. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| GTP binding | Binding to GTP, guanosine triphosphate. |
| GTPase activity | Catalysis of the reaction: GTP + H2O = GDP + H+ + phosphate. |
2 GO annotations of biological process
| Name | Definition |
|---|---|
| autophagosome assembly | The formation of a double membrane-bounded structure, the autophagosome, that occurs when a specialized membrane sac, called the isolation membrane, starts to enclose a portion of the cytoplasm. |
| intracellular protein transport | The directed movement of proteins in a cell, including the movement of proteins between specific compartments or structures within a cell, such as organelles of a eukaryotic cell. |
13 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q3ZC27 | RAB19 | Ras-related protein Rab-19 | Bos taurus (Bovine) | PR |
| Q5ZHV1 | RAB33B | Ras-related protein Rab-33B | Gallus gallus (Chicken) | PR |
| Q9H082 | RAB33B | Ras-related protein Rab-33B | Homo sapiens (Human) | PR |
| Q9NRW1 | RAB6B | Ras-related protein Rab-6B | Homo sapiens (Human) | PR |
| Q5JT25 | RAB41 | Ras-related protein Rab-41 | Homo sapiens (Human) | PR |
| P20340 | RAB6A | Ras-related protein Rab-6A | Homo sapiens (Human) | PR |
| Q9NP72 | RAB18 | Ras-related protein Rab-18 | Homo sapiens (Human) | PR |
| Q9ULC3 | RAB23 | Ras-related protein Rab-23 | Homo sapiens (Human) | PR |
| P20339 | RAB5A | Ras-related protein Rab-5A | Homo sapiens (Human) | PR |
| O35963 | Rab33b | Ras-related protein Rab-33B | Mus musculus (Mouse) | PR |
| P35294 | Rab19 | Ras-related protein Rab-19 | Mus musculus (Mouse) | PR |
| Q5M7U5 | Rab19 | Ras-related protein Rab-19 | Rattus norvegicus (Rat) | PR |
| Q20365 | rab-33 | Ras-related protein Rab-33 | Caenorhabditis elegans | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MHFSSSARAA | DENFDYLFKI | ILIGDSNVGK | TCVVQHFKSG | VYTETQQNTI | GVDFTVRSLD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| IDGKKVKMQV | WDTAGQERFR | TITQSYYRSA | HAAIIAYDLT | RRSTFESIPH | WIHEIEKYGA |
| 130 | 140 | 150 | 160 | 170 | 180 |
| ANVVIMLIGN | KCDLWEKRHV | LFEDACTLAE | KYGLLAVLET | SAKESKNIEE | VFVLMAKELI |
| 190 | 200 | 210 | |||
| ARNSLHLYGE | SALNGLPLDS | SPVLMAQGPS | EKTHCTC |