A3KGF7
Gene name |
Plcb2 |
Protein name |
1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 |
Names |
Phosphoinositide phospholipase C-beta-2, Phospholipase C-beta-2, PLC-beta-2 |
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:18796 |
EC number |
3.1.4.11: Phosphoric diester hydrolases |
Protein Class |
|
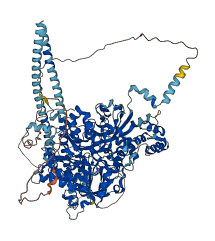
Descriptions
(Annotation based on sequence homology with Q9NQ66)
PLCB1 catalyzes the hydrolysis of 1-phosphatidylinositol 4,5-bisphosphate into diacylglycerol (DAG) and inositol 1,4,5-trisphosphate (IP3), and mediates intracellular signaling downstream of G protein-coupled receptors. Autoinhibitory region is a portion of the linker that separates the conserved X and Y boxes comprising the catalytic TIM barrel, at residues 468-539, and occludes the active site of PLCB1. The active site of a PLC isozyme toward phospholipid membranes should force the negatively charged X/Y linker away from the active site, therein relieving autoinhibition of the enzyme.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for A3KGF7
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A3KGF7-F1 | Predicted | AlphaFoldDB |
65 variants for A3KGF7
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs3388579696 | 27 | D>E | No | EVA | |
| rs3388578596 | 30 | T>I | No | EVA | |
| rs3388577540 | 37 | I>N | No | EVA | |
| rs3388581733 | 40 | V>E | No | EVA | |
| rs3388568061 | 41 | D>E | No | EVA | |
| rs3388579428 | 42 | P>H | No | EVA | |
| rs216938738 | 51 | Y>H | No | EVA | |
| rs3388575017 | 64 | I>S | No | EVA | |
| rs3388578602 | 77 | K>E | No | EVA | |
| rs3388577482 | 99 | L>P | No | EVA | |
| rs3388577504 | 130 | V>L | No | EVA | |
| rs3388575048 | 148 | L>M | No | EVA | |
| rs3388574988 | 175 | P>H | No | EVA | |
| rs3388579479 | 196 | D>Y | No | EVA | |
| rs3388580061 | 217 | P>H | No | EVA | |
| rs256817932 | 230 | S>A | No | EVA | |
| rs3388580093 | 230 | S>Y | No | EVA | |
| rs3388580011 | 248 | K>N | No | EVA | |
| rs3388573472 | 285 | Q>R | No | EVA | |
| rs3388581719 | 312 | Q>* | No | EVA | |
| rs3388578562 | 355 | C>* | No | EVA | |
| rs3388568077 | 404 | I>T | No | EVA | |
| rs223878077 | 426 | S>T | No | EVA | |
| rs3388574975 | 477 | P>T | No | EVA | |
| rs3388568034 | 505 | V>M | No | EVA | |
| rs3388568073 | 508 | E>D | No | EVA | |
| rs3388568036 | 543 | A>T | No | EVA | |
| rs3388577512 | 564 | F>I | No | EVA | |
| rs3388573718 | 577 | F>I | No | EVA | |
| rs3388579475 | 628 | M>K | No | EVA | |
| rs3388578549 | 633 | F>I | No | EVA | |
| rs3388581697 | 663 | R>C | No | EVA | |
| rs3388581671 | 703 | Y>F | No | EVA | |
| rs3388583020 | 767 | R>P | No | EVA | |
| rs3144311 | 790 | A>P | No | EVA | |
| rs3388568054 | 805 | Y>* | No | EVA | |
| rs3388582431 | 820 | P>S | No | EVA | |
| rs3388575022 | 864 | N>D | No | EVA | |
| rs3149505 | 867 | T>A | No | EVA | |
| rs3388568052 | 871 | T>A | No | EVA | |
| rs3388578927 | 873 | A>D | No | EVA | |
| rs3388582439 | 875 | G>R | No | EVA | |
| rs3388578554 | 892 | E>V | No | EVA | |
| rs3144309 | 894 | R>W | No | EVA | |
| rs3388575409 | 896 | L>M | No | EVA | |
| rs3388579635 | 918 | A>T | No | EVA | |
| rs3388580014 | 931 | Q>K | No | EVA | |
| rs49552524 | 938 | Q>P | No | EVA | |
| rs264382155 | 940 | A>S | No | EVA | |
| rs3388578865 | 941 | G>C | No | EVA | |
| rs212164187 | 971 | R>G | No | EVA | |
| rs3388573715 | 974 | P>L | No | EVA | |
| rs3144307 | 1007 | T>I | No | EVA | |
| rs3388571704 | 1009 | Q>P | No | EVA | |
| rs3388578869 | 1021 | Q>L | No | EVA | |
| rs255745385 | 1058 | T>I | No | EVA | |
| rs3388573727 | 1064 | Q>R | No | EVA | |
| rs3388577530 | 1086 | M>L | No | EVA | |
| rs3392122257 | 1088 | E>* | No | EVA | |
| rs3388573729 | 1093 | H>Q | No | EVA | |
| rs3388578566 | 1094 | Q>R | No | EVA | |
| rs3388578590 | 1135 | K>N | No | EVA | |
| rs3144294 | 1149 | E>K | No | EVA | |
| rs3388568075 | 1152 | D>G | No | EVA | |
| rs3388568055 | 1160 | A>V | No | EVA |
No associated diseases with A3KGF7
8 regional properties for A3KGF7
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | C2 domain | 666 - 791 | IPR000008 |
| domain | Phosphatidylinositol-specific phospholipase C, X domain | 312 - 464 | IPR000909 |
| domain | Phospholipase C, phosphatidylinositol-specific, Y domain | 547 - 663 | IPR001711 |
| domain | Phospholipase C-beta, C-terminal domain | 975 - 1149 | IPR014815 |
| domain | Phosphoinositide-specific phospholipase C, EF-hand-like domain | 214 - 303 | IPR015359 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2, catalytic domain | 311 - 650 | IPR028403 |
| domain | PLC-beta, PH domain | 13 - 144 | IPR037862 |
| domain | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2, EF-hand domain | 149 - 299 | IPR046969 |
Functions
| Description | ||
|---|---|---|
| EC Number | 3.1.4.11 | Phosphoric diester hydrolases |
| Subcellular Localization |
|
|
| PANTHER Family | ||
| PANTHER Subfamily | ||
| PANTHER Protein Class | ||
| PANTHER Pathway Category | No pathway information available | |
5 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytoplasm | The contents of a cell excluding the plasma membrane and nucleus, but including other subcellular structures. |
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| G-protein beta/gamma-subunit complex | The heterodimer formed by the beta and gamma subunits of a heterotrimeric G protein, which dissociates from the alpha subunit upon guanine nuclotide exchange. |
| Golgi apparatus | A membrane-bound cytoplasmic organelle of the endomembrane system that further processes the core oligosaccharides (e.g. N-glycans) added to proteins in the endoplasmic reticulum and packages them into membrane-bound vesicles. The Golgi apparatus operates at the intersection of the secretory, lysosomal, and endocytic pathways. |
| plasma membrane | The membrane surrounding a cell that separates the cell from its external environment. It consists of a phospholipid bilayer and associated proteins. |
5 GO annotations of molecular function
| Name | Definition |
|---|---|
| calcium ion binding | Binding to a calcium ion (Ca2+). |
| G-protein beta/gamma-subunit complex binding | Binding to a complex of G-protein beta/gamma subunits. |
| phosphatidylinositol phospholipase C activity | Catalysis of the reaction: 1-phosphatidyl-1D-myo-inositol 4,5-bisphosphate + H(2)O = 1,2-diacylglycerol + 1D-myo-inositol 1,4,5-trisphosphate + H(+). |
| phospholipase C activity | Catalysis of the reaction: a phospholipid + H2O = 1,2-diacylglycerol + a phosphatidate. |
| phospholipid binding | Binding to a phospholipid, a class of lipids containing phosphoric acid as a mono- or diester. |
6 GO annotations of biological process
| Name | Definition |
|---|---|
| detection of chemical stimulus involved in sensory perception of bitter taste | The series of events required for a bitter taste stimulus to be received and converted to a molecular signal. |
| lipid catabolic process | The chemical reactions and pathways resulting in the breakdown of lipids, compounds soluble in an organic solvent but not, or sparingly, in an aqueous solvent. |
| phosphatidylinositol metabolic process | The chemical reactions and pathways involving phosphatidylinositol, any glycophospholipid in which a sn-glycerol 3-phosphate residue is esterified to the 1-hydroxyl group of 1D-myo-inositol. |
| phosphatidylinositol-mediated signaling | The series of molecular signals in which a cell uses a phosphatidylinositol-mediated signaling to convert a signal into a response. Phosphatidylinositols include phosphatidylinositol (PtdIns) and its phosphorylated derivatives. |
| phospholipase C-activating G protein-coupled receptor signaling pathway | A G protein-coupled receptor signaling pathway in which the signal is transmitted via the activation of phospholipase C (PLC) and a subsequent increase in the intracellular concentration of inositol trisphosphate (IP3) and diacylglycerol (DAG). |
| sensory perception of bitter taste | The series of events required to receive a bitter taste stimulus, convert it to a molecular signal, and recognize and characterize the signal. This is a neurological process. |
24 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| P32383 | PLC1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase 1 | Saccharomyces cerevisiae (strain ATCC 204508 / S288c) (Baker's yeast) | PR |
| P10894 | PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Bos taurus (Bovine) | SS |
| Q9NQ66 | PLCB1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Homo sapiens (Human) | EV |
| Q15147 | PLCB4 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-4 | Homo sapiens (Human) | PR |
| Q01970 | PLCB3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | Homo sapiens (Human) | EV |
| Q00722 | PLCB2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | Homo sapiens (Human) | EV |
| Q8K394 | Plcl2 | Inactive phospholipase C-like protein 2 | Mus musculus (Mouse) | PR |
| Q8K2J0 | Plcd3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-3 | Mus musculus (Mouse) | PR |
| Q62077 | Plcg1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase gamma-1 | Mus musculus (Mouse) | SS |
| P51432 | Plcb3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | Mus musculus (Mouse) | SS |
| Q9Z1B3 | Plcb1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Mus musculus (Mouse) | SS |
| Q8K4S1 | Plce1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase epsilon-1 | Mus musculus (Mouse) | SS |
| Q8R3B1 | Plcd1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase delta-1 | Mus musculus (Mouse) | SS |
| P10687 | Plcb1 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-1 | Rattus norvegicus (Rat) | SS |
| Q99JE6 | Plcb3 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3 | Rattus norvegicus (Rat) | SS |
| O89040 | Plcb2 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-2 | Rattus norvegicus (Rat) | PR |
| G5EBH0 | egl-8 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta egl-8 | Caenorhabditis elegans | SS |
| Q8GV43 | PLC6 | Phosphoinositide phospholipase C 6 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944C2 | PLC5 | Phosphoinositide phospholipase C 5 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q6NMA7 | PLC9 | Phosphoinositide phospholipase C 9 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q9STZ3 | PLC8 | Phosphoinositide phospholipase C 8 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q56W08 | PLC3 | Phosphoinositide phospholipase C 3 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q39032 | PLC1 | Phosphoinositide phospholipase C 1 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| Q944C1 | PLC4 | Phosphoinositide phospholipase C 4 | Arabidopsis thaliana (Mouse-ear cress) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MSLLNPVLLP | PNVKAYLSQG | ERFIKWDDET | SIASPVILRV | DPKGYYLYWT | YQNQEMEFLD |
| 70 | 80 | 90 | 100 | 110 | 120 |
| VTSIRDTRFG | KFAKIPKSQK | LREVFNMDFP | DNHFLLKTLT | VVSGPDMVDL | TFYNFVSYKE |
| 130 | 140 | 150 | 160 | 170 | 180 |
| NVGKDWAEDV | LALAKHPMTV | NAPRSTFLDK | ILVKLKMQLN | PEGKIPVKNF | FQMFPADRKR |
| 190 | 200 | 210 | 220 | 230 | 240 |
| VEAALGACHL | AKGKNDAINP | EDFPESVYKS | FLMSLCPRPE | IDEIFTSYHS | KAKPYMTKEH |
| 250 | 260 | 270 | 280 | 290 | 300 |
| LTKFINQKQR | DPRLNSLLFP | PARPEQVQVL | IDKYEPSGIN | VQRGQLSPEG | MVWFLCGPEN |
| 310 | 320 | 330 | 340 | 350 | 360 |
| SVLAHDTLLI | HQDMTQPLNH | YFINSSHNTY | LTAGQFSGLS | SAEMYRQVLL | SGCRCVELDC |
| 370 | 380 | 390 | 400 | 410 | 420 |
| WKGKPPDEEP | IITHGFTMTT | DILFKEAIEA | IAESAFKTSP | YPVILSFENH | VDSPRQQAKM |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AEYCRSMFGE | TLLTDPLENF | PLKPGIPLPS | PEDLRGKILI | KNKKNQFSGP | ASPSKKPGGV |
| 490 | 500 | 510 | 520 | 530 | 540 |
| AEGSLPSSVP | VEEDTGWTAE | DRTEVEEEEV | VEEEEEEESG | NLDEEEIKKM | QSDEGTAGLE |
| 550 | 560 | 570 | 580 | 590 | 600 |
| VTAYEEMSSL | VNYIQPTKFI | SFEFSAQKNR | SYVVSSFTEL | KAYELLSKAS | MQFVDYNKRQ |
| 610 | 620 | 630 | 640 | 650 | 660 |
| MSRVYPKGTR | MDSSNYMPQM | FWNAGCQMVA | LNFQTMDLPM | QQNMALFEFN | GQSGYLLKHE |
| 670 | 680 | 690 | 700 | 710 | 720 |
| FMRRLDKQFN | PFSVDRIDVV | VATTLSITII | SGQFLSERSV | RTYVEVELFG | LPGDPKRRYR |
| 730 | 740 | 750 | 760 | 770 | 780 |
| TKLSPTANSI | NPVWKEEPFI | FEKILMPELA | SLRIAVMEEG | SKFLGHRIIP | INALHSGYHH |
| 790 | 800 | 810 | 820 | 830 | 840 |
| LCLRSESNMA | LTMPALFVFL | EMKDYIPDTW | ADLTVALANP | IKYFNAQDKK | SVKLKGVTGS |
| 850 | 860 | 870 | 880 | 890 | 900 |
| LPEKLFSGTP | VASQSNGAPV | SAGNGSTAPG | TKATGEEATK | EVTEPQTASL | EELRELKGVV |
| 910 | 920 | 930 | 940 | 950 | 960 |
| KLQRRHEKEL | RELERRGARR | WEELLQRGAA | QLAELQTQAA | GCKLRPGKGS | RKKRTLPCEE |
| 970 | 980 | 990 | 1000 | 1010 | 1020 |
| TVVAPSEPHD | RADPRVQELK | DRLEQELQQQ | GEEQYRSVLK | RKEQHVTEQI | AKMMELAREK |
| 1030 | 1040 | 1050 | 1060 | 1070 | 1080 |
| QAAELKTFKE | TSETDTKEMK | KKLEAKRLER | IQAMTKVTTD | KVAQERLKRE | INNSHIQEVV |
| 1090 | 1100 | 1110 | 1120 | 1130 | 1140 |
| QAVKQMTETL | ERHQEKLEER | QTACLEQIQA | MEKQFQEKAL | AEYEAKMKGL | EAEVKESVRA |
| 1150 | 1160 | 1170 | 1180 | ||
| YFKDCFPTEA | EDKPERSCEA | SEESCPQEPL | VSKADTQESR | L |