A2AGX3
Gene name |
Prdm11 |
Protein name |
PR domain-containing protein 11 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
mmu:100042784 |
EC number |
|
Protein Class |
|
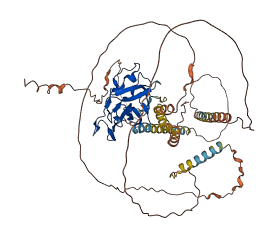
Descriptions
The autoinhibited protein was predicted that may have potential autoinhibitory elements via cis-regPred.
Autoinhibitory domains (AIDs)
Target domain |
|
Relief mechanism |
|
Assay |
cis-regPred |
Accessory elements
No accessory elements
Autoinhibited structure

Activated structure

1 structures for A2AGX3
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A2AGX3-F1 | Predicted | AlphaFoldDB |
32 variants for A2AGX3
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs250298317 | 48 | S>F | No | EVA | |
| rs864307050 | 57 | K>M | No | EVA | |
| rs3388571283 | 201 | F>Y | No | EVA | |
| rs3388572800 | 205 | E>D | No | EVA | |
| rs3388571308 | 220 | R>W | No | EVA | |
| rs3388567248 | 221 | L>W | No | EVA | |
| rs3388572780 | 237 | S>P | No | EVA | |
| rs3388571304 | 243 | R>C | No | EVA | |
| rs27382528 | 261 | L>M | No | EVA | |
| rs218172610 | 262 | E>D | No | EVA | |
| rs3388571354 | 272 | Q>R | No | EVA | |
| rs226739535 | 292 | I>M | No | EVA | |
| rs3388566714 | 293 | H>D | No | EVA | |
| rs265244480 | 296 | A>T | No | EVA | |
| rs3388562695 | 309 | L>P | No | EVA | |
| rs3388572798 | 329 | L>R | No | EVA | |
| rs3388574774 | 341 | D>E | No | EVA | |
| rs3388575593 | 356 | G>R | No | EVA | |
| rs3388564474 | 380 | P>H | No | EVA | |
| rs3388574819 | 413 | K>T | No | EVA | |
| rs3388576788 | 432 | P>S | No | EVA | |
| rs222324653 | 439 | P>A | No | EVA | |
| rs3388573613 | 451 | A>E | No | EVA | |
| rs3388572802 | 470 | R>H | No | EVA | |
| rs33064050 | 484 | S>L | No | EVA | |
| rs27382533 | 484 | S>P | No | EVA | |
| rs578764055 | 485 | Q>P | No | EVA | |
| rs3388574786 | 511 | V>L | No | EVA | |
| rs3388573612 | 536 | N>D | No | EVA | |
| rs3388564467 | 537 | K>I | No | EVA | |
| rs247595077 | 542 | C>R | No | EVA | |
| rs228575157 | 542 | C>S | No | EVA |
No associated diseases with A2AGX3
2 GO annotations of cellular component
| Name | Definition |
|---|---|
| cytosol | The part of the cytoplasm that does not contain organelles but which does contain other particulate matter, such as protein complexes. |
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| chromatin binding | Binding to chromatin, the network of fibers of DNA, protein, and sometimes RNA, that make up the chromosomes of the eukaryotic nucleus during interphase. |
| methyltransferase activity | Catalysis of the transfer of a methyl group to an acceptor molecule. |
8 GO annotations of biological process
| Name | Definition |
|---|---|
| methylation | The process in which a methyl group is covalently attached to a molecule. |
| negative regulation of cell growth | Any process that stops, prevents, or reduces the frequency, rate, extent or direction of cell growth. |
| negative regulation of DNA-templated transcription | Any process that stops, prevents, or reduces the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of DNA-templated transcription | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
| positive regulation of fibroblast apoptotic process | Any process that activates or increases the frequency, rate or extent of fibroblast apoptotic process. |
| regulation of cell cycle | Any process that modulates the rate or extent of progression through the cell cycle. |
| regulation of gene expression | Any process that modulates the frequency, rate or extent of gene expression. Gene expression is the process in which a gene's coding sequence is converted into a mature gene product (protein or RNA). |
| regulation of MAPK cascade | Any process that modulates the frequency, rate or extent of signal transduction mediated by the MAP kinase (MAPK) cascade. |
4 homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| Q9NQX1 | PRDM5 | PR domain zinc finger protein 5 | Homo sapiens (Human) | PR |
| Q9NQX0 | PRDM6 | Putative histone-lysine N-methyltransferase PRDM6 | Homo sapiens (Human) | PR |
| Q9CXE0 | Prdm5 | PR domain zinc finger protein 5 | Mus musculus (Mouse) | PR |
| Q60636 | Prdm1 | PR domain zinc finger protein 1 | Mus musculus (Mouse) | PR |
| 10 | 20 | 30 | 40 | 50 | 60 |
| MTENMKECLA | HTKAAVGDMV | TVVKTEVCSP | LRDQEYGQPC | SRRLEPSSME | VEPKKLKGKR |
| 70 | 80 | 90 | 100 | 110 | 120 |
| DLIVTKSFQQ | VDFWFCESCQ | EYFVDECPNH | GPPVFVSDTP | VPVGIPDRAA | LTIPQGMEVV |
| 130 | 140 | 150 | 160 | 170 | 180 |
| KDAGGESDVR | CINEVIPKGH | IFGPYEGQIS | TQDKSAGFFS | WLIVDKNNRY | KSIDGSDETK |
| 190 | 200 | 210 | 220 | 230 | 240 |
| ANWMRYVVIS | REEREQNLLA | FQHSERIYFR | ACRDIRPGER | LRVWYSEDYM | KRLHSMSQET |
| 250 | 260 | 270 | 280 | 290 | 300 |
| IHRNLARGEK | RLQREKAEQA | LENPEDLRGP | TQFPVLKQGR | SPYKRSFDEG | DIHPQAKKKK |
| 310 | 320 | 330 | 340 | 350 | 360 |
| IDLIFKDVLE | ASLESGNVEA | RQLALSTSLV | IRKVPKYQDD | DYGRAALTQG | ICRTPGEGDW |
| 370 | 380 | 390 | 400 | 410 | 420 |
| KVPQRVAKEL | GPLEDEEEEP | TSFKADSPAE | ASLASDPHEL | PTTSFCPNCI | RLKKKVRELQ |
| 430 | 440 | 450 | 460 | 470 | 480 |
| AELDMLKSGK | LPEPSLLPPQ | VLELPEFSDP | AGKFLRMRLL | LKGRVCSATR | AHCVEGGPER |
| 490 | 500 | 510 | 520 | 530 | 540 |
| SALSQPPARR | PEECNSAEAA | PSWGGSPVPQ | VTEAGWLEDR | GREGIACANL | YILLVNKICP |
| 550 | 560 | ||||
| ICRRAVCCVL | DLVFSLGGGA | HCCLP |