A2A5C2
Gene name |
Etv4 |
Protein name |
ETS translocation variant 4 |
Names |
|
Species |
Mus musculus (Mouse) |
KEGG Pathway |
|
EC number |
|
Protein Class |
ETS (PTHR11849) |
Descriptions
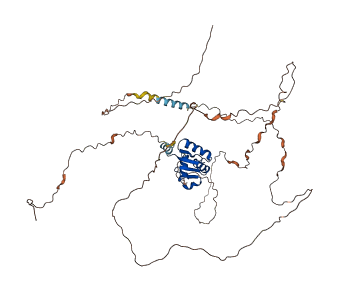
Etv4 is a transcriptional activator playing a role in keratinocyte differentiation. Truncation of the autoinhibitory regions increases the transcriptional and DNA binding activities of Etv4. The autoinhibition of Etv4 is mediated by elements outside of ETS domain, and exists in two conformations: a closed, inhibited form and an open, active form that is competent to bind to DNA.
Autoinhibitory domains (AIDs)
Target domain |
334-417 (ETS domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay |
Target domain |
334-417 (ETS domain) |
Relief mechanism |
Partner binding |
Assay |
Deletion assay, Mutagenesis experiment |
Accessory elements
No accessory elements
References
- Bojović BB et al. (2001) "The PEA3 Ets transcription factor comprises multiple domains that regulate transactivation and DNA binding", The Journal of biological chemistry, 276, 4509-21
- Green SM et al. (2010) "DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association", The Journal of biological chemistry, 285, 18496-504
Autoinhibited structure

Activated structure

1 structures for A2A5C2
| Entry ID | Method | Resolution | Chain | Position | Source |
|---|---|---|---|---|---|
| AF-A2A5C2-F1 | Predicted | AlphaFoldDB |
5 variants for A2A5C2
| Variant ID(s) | Position | Change | Description | Diseaes Association | Provenance |
|---|---|---|---|---|---|
| rs260245572 | 66 | A>V | No | Ensembl | |
| rs235382712 | 103 | R>C | No | Ensembl | |
| rs234101126 | 220 | N>S | No | Ensembl | |
| rs28234792 | 322 | I>F | No | Ensembl | |
| rs28234799 | 473 | G>S | No | Ensembl |
No associated diseases with A2A5C2
1 regional properties for A2A5C2
| Type | Name | Position | InterPro Accession |
|---|---|---|---|
| domain | Ets domain | 336 - 421 | IPR000418 |
Functions
| Description | ||
|---|---|---|
| EC Number | ||
| Subcellular Localization |
|
|
| PANTHER Family | PTHR11849 | ETS |
| PANTHER Subfamily | PTHR11849:SF181 | ETS TRANSLOCATION VARIANT 4 |
| PANTHER Protein Class |
DNA-binding transcription factor
helix-turn-helix transcription factor winged helix/forkhead transcription factor |
|
| PANTHER Pathway Category | No pathway information available | |
1 GO annotations of cellular component
| Name | Definition |
|---|---|
| nucleus | A membrane-bounded organelle of eukaryotic cells in which chromosomes are housed and replicated. In most cells, the nucleus contains all of the cell's chromosomes except the organellar chromosomes, and is the site of RNA synthesis and processing. In some species, or in specialized cell types, RNA metabolism or DNA replication may be absent. |
2 GO annotations of molecular function
| Name | Definition |
|---|---|
| DNA-binding transcription factor activity | A transcription regulator activity that modulates transcription of gene sets via selective and non-covalent binding to a specific double-stranded genomic DNA sequence (sometimes referred to as a motif) within a cis-regulatory region. Regulatory regions include promoters (proximal and distal) and enhancers. Genes are transcriptional units, and include bacterial operons. |
| sequence-specific DNA binding | Binding to DNA of a specific nucleotide composition, e.g. GC-rich DNA binding, or with a specific sequence motif or type of DNA e.g. promotor binding or rDNA binding. |
1 GO annotations of biological process
| Name | Definition |
|---|---|
| positive regulation of transcription, DNA-templated | Any process that activates or increases the frequency, rate or extent of cellular DNA-templated transcription. |
No homologous proteins in AiPD
| UniProt AC | Gene Name | Protein Name | Species | Evidence Code |
|---|---|---|---|---|
| No homologous proteins | ||||
| 10 | 20 | 30 | 40 | 50 | 60 |
| MERRMKGGYL | DQRVPYTFCS | KSPGNGSLGE | ALMVPQGKLM | DPGSLPPSDS | EDLFQDLSHF |
| 70 | 80 | 90 | 100 | 110 | 120 |
| QETWLAEAQV | PDSDEQFVPD | FHSENLAFHS | PTTRIKKEPQ | SPRTDPALSC | SRKPPLPYHH |
| 130 | 140 | 150 | 160 | 170 | 180 |
| GEQCLYSRQI | AIKSPAPGAP | GQSPLQPFSR | AEQQQSLLRA | SSSSQSHPGH | GYLGEHSSVF |
| 190 | 200 | 210 | 220 | 230 | 240 |
| QQPVDMCHSF | TSPQGGGREP | LPAPYQHQLS | EPCPPYPQQN | FKQEYHDPLY | EQAGQPASSQ |
| 250 | 260 | 270 | 280 | 290 | 300 |
| GGVSGHRYPG | AGVVIKQERT | DFAYDSDVPG | CASMYLHPEG | FSGPSPGDGV | MGYGYEKSLR |
| 310 | 320 | 330 | 340 | 350 | 360 |
| PFPDDVCIVP | EKFEGDIKQE | GIGAFREGPP | YQRRGALQLW | QFLVALLDDP | TNAHFIAWTG |
| 370 | 380 | 390 | 400 | 410 | 420 |
| RGMEFKLIEP | EEVARLWGIQ | KNRPAMNYDK | LSRSLRYYYE | KGIMQKVAGE | RYVYKFVCEP |
| 430 | 440 | 450 | 460 | 470 | |
| EALFSLAFPD | NQRPALKAEF | DRPVSEEDTV | PLSHLDESPA | YLPELTGPAP | PFGHRGGYSY |